Abstract
Microdochium species have often been reported as plant pathogens and saprophytes and are commonly isolated from some diseased plant hosts. The primary aim of the present study was to describe and illustrate two new Microdochium species isolated from the leaf spot of Indocalamus longiauritus in Yunnan Province, China, namely Microdochium yunnanense and M. indocalami, spp. nov., based on their morphology and multilocus phylogenetic analyses of the combined ITS, LSU, TUB2, and RPB2. DNA sequence data indicate that six strains represent three independent groups from related and similar species in Microdochium. Microdochium indocalamisp. nov. clustered with M. fisheri, M. lycopodinum, M. rhopalostylidis, and M. phragmitis. Microdochium yunnanensesp. nov. grouped with M. bolleyi. In addition, the strain SAUCC1017 is recorded as an unidentified species in Microdochium. Descriptions and illustrations of the new species in the genus and Microdochium sp. indet. are provided.
Keywords: Microdochiaceae , multigene phylogeny, new species, taxonomy, Xylariales
Introduction
Microdochium is a genus in Microdochiaceae (Xylariales), which has been well-studied in recent years by Hernández-Restrepo et al. (2016), Zhang et al. (2017), Crous et al. (2018, 2019), and Marin-Felix et al. (2019) by incorporating morphological and molecular data with appropriate genes to resolve species limitations in the genus. Microdochium phragmitis, the type of the genus, was introduced by Sydow (1924) for an ascomycetous fungal plant pathogen found on leaves of Phragmites australis (= Phragmites communis) in Germany in 1919, which has globose, erumpent stromata of minute, hyaline cells, small papillate conoid conidiogenous cells, and solitary, fusiform to subfalcate hyaline conidia. Currently, about 48 species of this genus are listed in Index Fungorum (http://www.indexfungorum.org/; accessed 1 May 2020), but only about two-fifths of them are well known and have been studied in pure culture (Crous et al. 2018, 2019; Marin-Felix et al. 2019).
Monographella was described by Petrak (1924) and was considered the sexual morph of Microdochium for many years (Hernández-Restrepo et al. 2016). Nevertheless, Microdochium has more species, is more commonly encountered, and the name is more frequently used in the literature. With the implementation of “one fungus one name” nomenclature, Microdochium has been retained as a genus name (Hernández-Restrepo et al. 2016).
Microdochium included important plant pathogens, particularly on grasses and cereals. Kwasna et al. (2007) newly described M. triticicola which was isolated from roots of wheat in the United Kingdom. Zhang et al. (2015) identified M. paspali, which caused leaf blight of seashore paspalum (Paspalum vaginatum), a turfgrass widely used in tropical and subtropical golf courses. Crous et al. (2018) described M. musae isolated from leaves of Musa sp. Microdochium rhopalostylidis, found on the leaves of Rhopalostylis sapida (Arecaceae) in New Zealand, was identified and described by Crous et al. (2019). From turf leaves (Poaceae) in New Zealand, M. novae-zelandiae was isolated by Marin-Felix et al. (2019) and described as a new species.
Many taxonomic problems have occurred in Microdochium, and the genus was shown to be polyphyletic (Hernández-Restrepo et al. 2016). However, some species have been reclassified based on molecular analyses (Glynn et al. 2005; Jewell and Hsiang 2013; Hernández-Restrepo et al. 2016). For example, phylogenetic analysis of the translation elongation factor 1-alpha gene (TEF1) showed that the isolates previously described as varieties of Microdochium nivale shown a distinct heterogeneity between isolates and these isolates were generated as two separate species, M. majus and M. nivale (Glynn et al. 2005). The study of multigene differences between M. nivale and M. majus by Jewell and Hsiang (2013) supported the reclassification of M. nivale and M. majus as sister species rather than varieties. Three species of Microdochium were revised by Hernández-Restrepo et al. (2016), which were initially recognised as M. gracile (CBS 493.70), M. tripsaci (CBS 857.72), and M. fusarioides (CBS 740.83, CBS 741.83, and CBS 742.83), and were renamed Paramicrodochium gracile (Sordariomycetesincertae sedis), Ephelis tripsaci (Clavicipitaceae, Hypocreales), and Microdochiella fusarioides (Orbiliales).
In this study, we introduce two novel species, M. yunnanense and M. indocalami spp. nov., which were isolated from the leaves of Indocalamus longiauritus in China. These two species are introduced based on both morphological features and molecular sequence data.
Materials and methods
Isolation and morphological studies
The samples were collected from Yunnan Province, China. The strains of Microdochium were isolated from diseased or healthy leaves of Indocalamus longiauritus using single spore and tissue isolation methods (Chomnunti et al. 2014). Single spore isolation following the protocol of Choi et al. (1999) and Zhang et al. (2013) was adopted for collection with visible foliar sporulation. The spore suspension was obtained and spread onto potato dextrose agar (PDA) and incubated overnight under normal conditions. The germinated spores were then transferred to a new PDA plate to obtain a pure culture. Besides, the surface-sterilised plant tissue isolation was also used to obtain sterile isolates from plant host. Fungi were isolated by cutting eight fragments (5 × 5 mm) per leaf from the margin of leaf lesions and surface-sterilized by consecutively immersing in 75% ethanol solution for 1 min, 5% sodium hypochlorite solution for 30 s, and then rinsing in sterile distilled water for 1 min (Gao et al. 2014; Liu et al. 2015). The samples were dried with sterilized paper towels and placed on potato dextrose agar (PDA) (Cai et al. 2009). All the plates were incubated at biochemical incubator at 25 °C for 3–4 days, then hyphae were picked out of the periphery of the colonies and inoculated onto new PDA plates.
Following 2–3 weeks of incubation, morphological characters were recorded as by Hernández-Restrepo et al. (2016). Photographs of the colonies were taken at 7 days and 15 days using a Powershot G7X mark II digital camera. Micromorphological characters were observed using an Olympus SZX10 stereomicroscope and an Olympus BX53 microscope, both fitted with Olympus DP80 high definition color digital cameras to photo-document fungal structures. All fungal strains were stored in 10% sterilized glycerin at 4 °C for further studies. Voucher specimens were deposited in the Herbarium of the Department of Plant Pathology, Shandong Agricultural University (HSAUP). Living cultures were deposited in the Shandong Agricultural University Culture Collection (SAUCC). Taxonomic information of the new taxa was submitted to MycoBank (http://www.mycobank.org).
DNA extraction and amplification
Genomic DNA was extracted from fungal mycelia grown on PDA, using a modified cetyltrimethylammonium bromide (CTAB) protocol as described in Guo et al. (2000). Four pairs of primers were adopted to amplify gene sequences (Hernández-Restrepo et al. 2016). The partial large subunit (LSU) rDNA, the internal transcribed spacer region with intervening 5.8S nrRNA gene (ITS), part of the beta-tubulin gene region (TUB2), and partial RNA polymerase II second largest subunit (RPB2) region were amplified and sequenced using primers pairs LR0R/LR5 (Vilgalys and Hester 1990), ITS4/ITS5 (White et al. 1990), Btub526F and Btub1332R (Jewell and Hsiang 2013), and RPB2-5F2/fRPB2-7cR (Liu et al. 1999; Sung et al. 2007), respectively.
PCR was performed using an Eppendorf Master Thermocycler (Hamburg, Germany). Amplification reactions were performed in a 25 μL reaction volume, which contained 12.5 μL Green Taq Mix (vazyme, Nanjing, China), 1 μL of each forward and reverse primer (10 μM) (Biosune, Shanghai, China), and 1 μL template genomic DNA in amplifier, and were adjusted with distilled deionized water to a total volume of 25 μL.
PCR parameters were as follows: 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at a suitable temperature for 30 s, extension at 72 °C for 1 min and a final elongation step at 72 °C for 10 min. Annealing temperature for each gene were 55 °C for ITS, 51 °C for LSU, 56 °C for RPB2 and 53 °C for TUB2. The PCR products were visualised on 1% agarose electrophoresis gel. Sequencing was done bi-directionally, conducted by the Biosune Company Limited (Shanghai, China). Consensus sequences were obtained using MEGA 7.0 (Kumar et al. 2016). All sequences generated in this study were deposited in GenBank (Table 1).
Table 1.
Specimens and GenBank accession numbers of DNA sequences used in this study.
| Species | Voucher | Host/Substrate | Country | GeneBank accession numbers | |||
|---|---|---|---|---|---|---|---|
| LSU | ITS | TUB2 | RPB2 | ||||
| Idriella lunata | CBS 204.56* | Root of Fragaria chiloensis | USA | KP858981 | KP859044 | – | – |
| Microdochium albescens | CBS 291.79 | On Oryza sativa | Ivory Coast | KP858932 | KP858996 | KP859059 | KP859105 |
| CBS 243.83 | Seed Oryza sativa | Unknown country | KP858930 | KP858994 | KP859057 | KP859103 | |
| M. bolleyi | CBS 540.92 | Root of Hordeum vulgare | Syria | KP858946 | KP859010 | KP859073 | KP859119 |
| M. citrinidiscum | CBS 109067* | Leaf of Eichhornia crassipes | Peru | KP858939 | KP859003 | KP859066 | KP859112 |
| M. chrysanthemoides | LC5363 = CGMCC3.17929* | Unnamed Karst Cave | China | KU746736 | KU746690 | KU746781 | – |
| LC5466 = CGMCC3.17930 | Unnamed Karst Cave | China | KU746735 | KU746689 | KU746782 | – | |
| M. colombiense | CBS 624.94* | On Musa sapientum | Colombia | KP858935 | KP858999 | KP859062 | KP859108 |
| M. fisheri | CBS 242.90* | Stem of Oryza sativa | UK | KP858951 | KP859015 | KP859079 | KP859124 |
| M. indocalami | SAUCC1016* | Leaves of Indocalamus longiauritus | China | MT199878 | MT199884 | MT435653 | MT510550 |
| M. lycopodinum | CBS 146.68 | Air sample | The Netherlands | KP858929 | KP858993 | KP859056 | KP859102 |
| CBS 109397 | On Phragmites australis | Germany | KP858940 | KP859004 | KP859067 | KP859113 | |
| CBS 109398 | On Phragmites australis | Germany | KP858941 | KP859005 | KP859068 | KP859114 | |
| M. majus | CBS 741.79 | On Triticum aestivum | Germany | KP858937 | KP859001 | KP859064 | KP859110 |
| M. musae | CBS 111018 = CPC 5380 | Musa cv. Cavendish | Costa Rica | – | AY293061 | – | – |
| CBS 143499 = CPC 32809 | Leaves of Musa sp. | Malaysia | MH107941 | MH107894 | MH108040 | – | |
| CBS 143500* = CPC 32689 | Leaves of Musa sp. | Malaysia | MH107942 | MH107895 | MH108041 | MH108003 | |
| CPC 11234 | Leaves of Musa sp. | Mauritius | MH107943 | MH107896 | MH108042 | – | |
| CPC 11240 | Leaves of Musa sp. | Mauritius | MH107944 | MH107897 | MH108043 | – | |
| CPC 16258 | Leaves of Musa sp. | Mexico | MH107945 | MH107898 | MH108044 | – | |
| CPC 32681 | Leaves of Musa sp. | Malaysia | MH107946 | MH107899 | – | – | |
| M. neoqueenslandicum | CBS 445.95 | On Juncus effusus | The Netherlands | KP858933 | KP858997 | KP859060 | KP859106 |
| CBS 108926* | On Agrostis sp. | New Zealand | KP858938 | KP859002 | KP859065 | KP859111 | |
| M. nivale | CBS 116205* | Roots of Triticum aestivum | UK | KP858944 | KP859008 | KP859071 | KP859117 |
| M. nivale var. nivale | CBS 288.50 | Unknown | Unknown country | MH868135 | MH856626 | – | – |
| M. novae-zelandiae | CBS 143847 | From turf leaves (Poaceae) | New Zealand | – | LT990655 | LT990608 | LT990641 |
| CPC 29693 | From turf leaves (Poaceae) | New Zealand | – | LT990656 | LT990609 | LT990642 | |
| M. paspali | HK-ML-1371 | Paspalum vaginatum | China | – | KJ569509 | KJ569514 | – |
| QH-BA-48 | Paspalum vaginatum | China | – | KJ569510 | KJ569515 | – | |
| SY-LQG66 | Paspalum vaginatum | China | – | KJ569511 | KJ569516 | – | |
| WC-WC-85 | Paspalum vaginatum | China | – | KJ569512 | KJ569517 | – | |
| WN-BD-452 | Paspalum vaginatum | China | – | KJ569513 | KJ569518 | – | |
| M. phragmitis | CBS 285.71* | On Phragmites australis | Poland | KP858949 | KP859013 | KP859077 | KP859122 |
| CBS 423.78 | On Phragmites communis | Germany | KP858948 | KP859012 | KP859076 | KP859121 | |
| M. rhopalostylidis | CPC 34449 = CBS 145125* | Rhopalostylis sapida | New Zealand | MK442532 | MK442592 | MK442735 | MK442667 |
| M. seminicola | KAS3576 = CBS 139951* | Maize kernels | Switzerland | KP858974 | KP859038 | KP859101 | KP859147 |
| KAS1516 = CPC 26001 | On grain | Canada | KP858961 | KP859025 | KP859088 | KP859134 | |
| KAS3574 = DAOM 250155 | Maize kernels | Switzerland | KP858973 | KP859037 | KP859100 | KP859146 | |
| KAS3158 = DAOM 250161 | On Triticum aestivum | Canada | KP858970 | KP859034 | KP859097 | KP859143 | |
| KAS1527 = DAOM 250165 | On grain | Canada | KP858966 | KP859030 | KP859093 | KP859139 | |
| KAS1473 = DAOM 250176 | On Triticum aestivum | Canada | KP858955 | KP859019 | KP859082 | KP859128 | |
| M. sorghi | CBS 691.96 | Living Sorghum halepense | Cuba | KP858936 | KP859000 | KP859063 | KP859109 |
| M. sp. indet. | SAUCC1017 | Leaves of Indocalamus longiauritus | China | MT199879 | MT199885 | MT435654 | – |
| M. tainanense | CBS 269.76* | Root of Saccharum officinarum | China, Taiwan | KP858945 | KP859009 | KP859072 | KP859118 |
| CBS 270.76 | Root of Saccharum officinarum | China, Taiwan | KP858931 | KP858995 | KP859058 | KP859104 | |
| M. trichocladiopsis | CBS 623.77* | Rhizosphere of Triticum aestivum | Unknown country | KP858934 | KP858998 | KP859061 | KP859107 |
| M. yunnanense | SAUCC1011* | Leaves of Indocalamus longiauritus | China | MT199875 | MT199881 | MT435650 | MT510547 |
| SAUCC1012 | Leaves of Indocalamus longiauritus | China | MT199876 | MT199882 | MT543651 | MT510548 | |
| SAUCC1015 | Leaves of Indocalamus longiauritus | China | MT199877 | MT199883 | MT435652 | MT510549 | |
| SAUCC1018 | Leaves of Indocalamus longiauritus | China | MT199880 | MT199886 | MT435655 | – | |
Isolates marked with “*” are ex-type or ex-epitype strain.
Phylogenetic analyses
Novel sequences generated from the six strains in this study, and all reference available sequences of Microdochium species downloaded from GenBank (mostly used in Hernández-Restrepo et al. 2016; Zhang et al. 2017; Marin-Felix et al. 2019; Crous et al. 2018, 2019) were used for phylogenetic analyses. Alignments of the individual locus were determined using MAFFT v. 7.110 by default settings (Katoh et al. 2017) and manually corrected where necessary. To establish the identity of the isolates at species level, phylogenetic analyses were conducted first individually for each locus and then as combined analyses of four loci (ITS, LSU, TUB2, and RPB2 regions). Phylogenetic analyses were based on maximum likelihood (ML) and Bayesian inference (BI) for the multi-locus analyses. For BI, the best evolutionary model for each partition was determined using MrModeltest v. 2.3 (Nylander 2004) and incorporated into the analyses. ML and BI were run on the CIPRES Science Gateway portal (https://www.phylo.org/) (Miller et al. 2012) using RaxML-HPC2 on XSEDE (8.2.12) (Stamatakis 2014) and MrBayes on XSEDE (3.2.7a) (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003; Ronquist et al. 2012), respectively. For ML analyses the default parameters were used and BI was carried out using the rapid bootstrapping algorithm with the automatic halt option. Bayesian analyses included four parallel runs of 5,000,000 generations, with the stop rule option and a sampling frequency of 500 generations. The burn-in fraction was set to 0.25 and posterior probabilities (PP) were determined from the remaining trees. The resulting trees were plotted using FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree) and edited with Adobe Illustrator CS5.1. New sequences generated in this study were deposited at GenBank (https://www.ncbi.nlm.nih.gov; Table 1), the alignments and trees were deposited in TreeBASE (http://treebase.org/treebase-web/home.html).
Results
Phylogenetic analyses
Six Microdochium strains isolated from plant hosts were sequenced. Microdochium was analysed by using multilocus data (ITS, LSU, TUB2 and RPB2) composed of 50 isolates of Microdochium and Idriella lunata (CBS 204.56) as the outgroup taxon. A total of 3257 characters including gaps were obtained in the phylogenetic analysis, viz. ITS: 1–572, LSU: 573–1429, TUB2: 1430–2395, RPB2: 2396–3257. Of these characters, 2019 were constant, 219 were variable and parsimony-uninformative, and 1019 were parsimony-informative. For the BI and ML analyses, GTR+I+G for LSU and RPB2, SYM+I+G for ITS, and GTR+G for TUB2 were selected and incorporated into the analyses. The ML tree topology confirmed the tree topologies obtained from the BI analyses, and therefore, only the ML tree is presented (Fig. 1).
Figure 1.
Phylogram of Microdochium based on combined ITS, LSU, TUB2 and RPB2 genes. The ML and BI bootstrap support values above 75% and 0.95 BYPP are shown at the first and second position, respectively. Strains marked with “*” are ex-type or ex-epitype. Strains from the current study are in red. Some branches were shortened to fit them to the page – these are indicated by two diagonal lines with the number of times a branch was shortened indicated next to the lines.
ML bootstrap support values (≥ 75%) and Bayesian posterior probability (≥ 0.95) are shown as first and second position above nodes, respectively. The 50 strains were assigned to 23 species clades based on the four gene loci phylogeny (Fig. 1). The six strains studied here represented two novel species. The new species of Microdochium yunnanense showed a close relationship to M. bolleyi (CBS 540.92) with good support (ML-BS: 98% and BYPP: 1.00). Microdochium indocalami (SAUCC1016) appeared most closely related to M. fisheri (CBS 242.90), M. lycopodinum (CBS 146.68), M. rhopalostylidis (CBS 145125), and M. phragmitis (CBS 285.71) with high support by the multi-locus phylogeny. From the tree (Fig. 1), strain SAUCC1017 formed a conspicuous branch independent from other Microdochium species, thus supporting the introduction of SAUCC1017 as an indeterminate species.
Taxonomy
Microdochium indocalami
S.T. Huang, J.W. Xia, X.G. Zhang, W.X. Sun & Z. Li sp. nov.
C2015F60-88EF-51A8-9D84-260F88607CFB
835766
Figure 2.
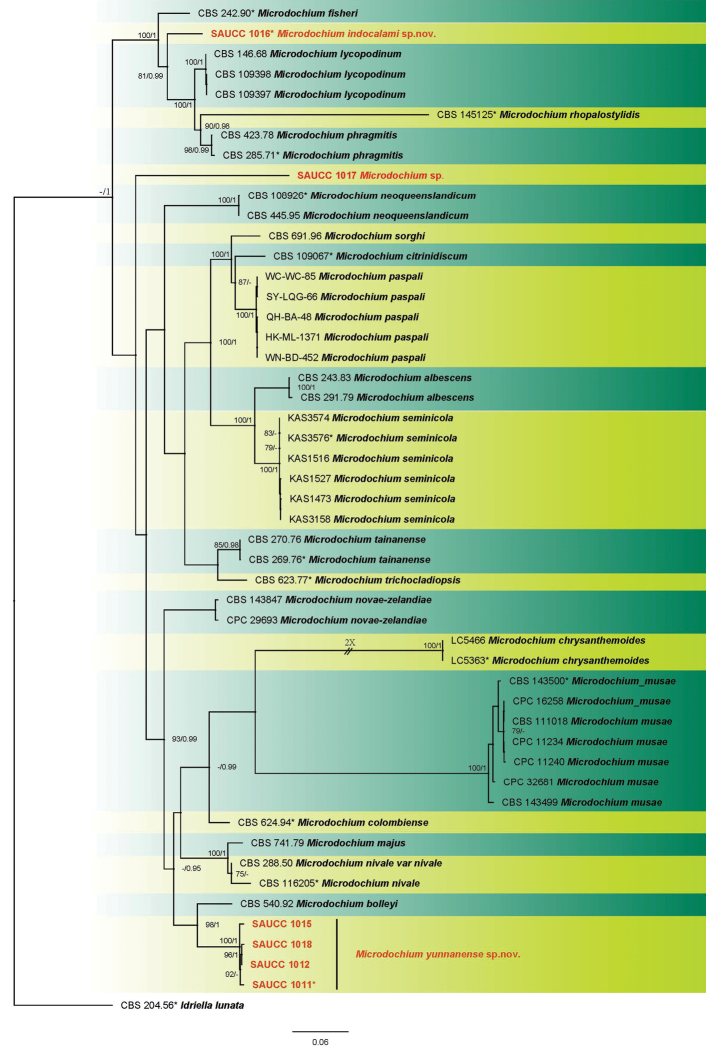
Microdochium indocalami (SAUCC1016) a leaves of host plant b colony overview c–e surface of colony after 15 days on PDA (c), OA (d), MEA (e) f–h reverse of colony after 15 days on PDA (f) OA (g) MEA (h) i–m conidiophores and conidiogenous cells n conidia. Scale bars: 10μm (i–n).
Etymology.
Name refers to the genus of the host plant Indocalamus longiauritus.
Diagnosis.
Characterised by the size of conidia and the number of septa of conidia.
Type.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Indocalamus longiauritus. 16 April 2019, S.T. Huang, HSAUP1016, holotype, ex-type living culture SAUCC1016.
Description.
Colonies on PDA attaining 46.1–51.2 mm in diameter after 7 days, formed a conspicuous concentric circle, periphery of aerial mycelium cottony, centre with scarce aerial mycelium, white initially, then becoming greyish sepia after 25 days. Some aerial hyphae aggregated and form a sporodochium within 15 days or longer. Mycelium composed of hyaline, immersed and superficial, smooth, branched, septate, 2.0–3.0 μm wide hyphae. Due to the soluble pigment secreted, reverse white to salmon. Conidiophores straight or slightly curved, aseptate, aggregated in the aerial mycelium, often reduced to conidiogenous cells borne directly from the hyphae. Conidiogenous cells terminal or intercalary, mono-or polyblastic, denticulate, smooth, hyaline, cylindrical, straight or bent, 11.0–28.3 × 1.5–2.9 μm. Conidia cylindrical, clavate to obovoid, 1–3-septate, 13.0–15.5 × 3.5–5.5 μm, base usually flattened 0.5–1.0 μm. Sometimes borne directly from the mycelial hyphae. Sexual morph: unknown.
Culture characteristics.
Colonies on OA 62.0–64.0 mm in diameter after 7 days, centre with aerial mycelium cottony, periphery with scarce aerial mycelium. Mycelium mostly immersed, hyphae hyaline, septate, smooth, exudate and soluble pigment produced, reverse white initially, then becoming pale mouse-grey in periphery and mouse-grey in center. Sporodochia formed on agar surface. Colonies on MEA 50.8–52.7 mm in diameter after 7 days, aerial mycelium abundant, with concentric rings, white to pale pink, periphery with cottony aerial mycelium, centre with scarce aerial mycelium, exudate absent. Reverse white to pale pink with age.
Habitat and distribution.
Isolated from leaves of Indocalamus longiauritus in China.
Notes.
Microdochium and allied genera were revised by Hernández-Restrepo et al. (2016). Phylogenetic analysis of a combined four gene showed that M. indocalami (strain SAUCC1016) formed a separated branch as the clade of M. fisheri, M. lycopodinum, M. phragmites, and M. rhopalostylidis with good support (ML-BS: 100% and BYPP: 1.00) (Fig. 1). Additionally, conidiogenous cells of M. indocalami are terminal or intercalary, denticulate, cylindrical which are similar to the species in this clade. The size of conidia and the number of septa of conidia reported for M. fisheri (7.0–12.0 × 3.0–4.0 μm, 0–1-septate), M. lycopodinum (8.0–15.5 × 2.5–4.0 μm, 0–1-septate), M. phragmites (10.0–14.5 × 2.0–3.0 μm, 0–1-septate), and M. rhopalostylidis ((13.0–) 16.0–20.0 (–23.0) × (2.5–) 3.0 (–4.0) μm, 1–3-septate) (Hernández-Restrepo et al. 2016; Crous et al. 2019) were different to M. indocalami (13.0–15.5 × 3.5–5.5 μm, 1–3-septate).
Microdochium yunnanense
S.T. Huang, J.W. Xia, X.G. Zhang, W.X. Sun & Z. Li sp. nov.
DA50595A-936A-5A65-AE44-C5BE833BC6D8
835765
Figure 3.
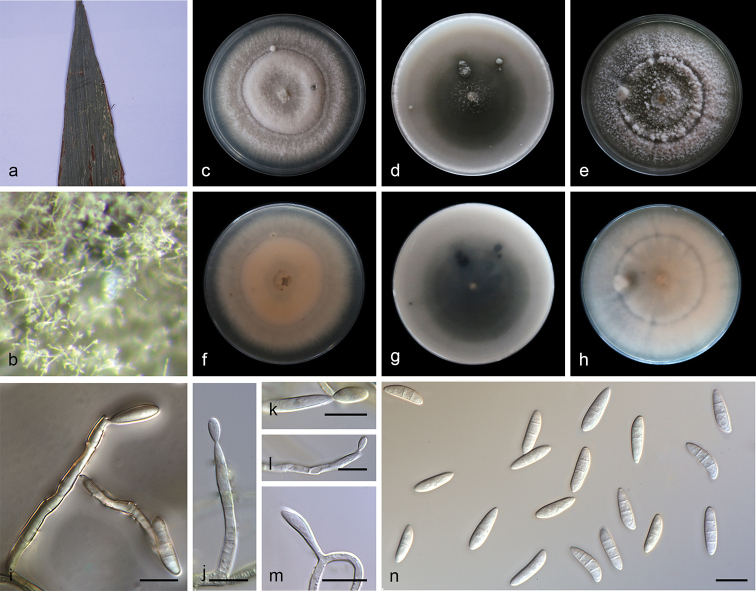
Microdochium yunnanense (SAUCC1011) a leaves of host plant b sporodochia on media surface c–e surface of colony after 15 days on PDA (c), OA (d), MEA (e) f–h reverse of colony after 15 days on PDA (f), OA (g), MEA (h) i sporodochial conidiophores and conidiogenous cells with conidia j conidiogenous cells k–l conidia. Scale bars: 10μm (i–l).
Etymology.
Named after Yunnan Province, where the fungus was collected.
Diagnosis.
Characterised by conidiomata sporodochium-like and the size of conidia.
Type.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Indocalamus longiauritus. 16 April 2019, S.T. Huang, HSAUP1011 holotype, ex-type living culture SAUCC1011.
Description.
Colonies on PDA attaining 48.0–61.5 mm in diameter after 15 days, felty, compact, erose or dentate, initially white, then becoming yellowish with age. Mycelium superficial, consisting of hyaline, smooth, branched, septate, 1.0–2.5 μm wide hyphae. Conidiomata sporodochium-like, appeared within 15 days or longer, formed in aerial mycelium or on agar surface, slimy, hyaline or orange, semi-submerged. Exudate occasionally appeared on old sporodochia. Reverse colorless to yellowish, due to the soluble pigment secreted. Conidiophores formed terminal or lateral with sympodial proliferation, solitary or aggregated. Most conidiophores tightly aggregated in a sporodochium, inconspicuous flat-tipped loci, irregularly branched, or borne directly on mycelial hyphae, straight or slightly curved, aseptate, guttulate, smooth-walled, apex subobtuse, base truncate. Conidiogenous cells of two types: some polyblastic, ampulliform, lageniform, with percurrent proliferations, 6.5–10.0 × 2.5–3.4 μm, neck up to 4.5 μm long, 1.0–1.5 μm wide, others straight or bent, smooth, cylindrical up to 10.0–11.5 μm long, 1.0–2.0 μm wide. Conidia aseptate, mostly lunate, a few ellipsoid and cylindrical, hyaline, straight or curved, obtuse, 0–2 guttulate in mature conidia, 6.8–10.0 × 2.4–3.5 μm. Chlamydospores was not observed. Sexual morph: unknown.
Culture characteristics.
Colonies on OA 58.1–61.5 mm in diameter after 15 days, entire, flat, white, lobate and radially margin, aerial mycelia cottony or sparse. Less exudate. Reverse white. Vegetative hyphae hyaline, abundant, branched, septate, thin-walled. Colonies on MEA 39.5–48.2 mm in diameter after 15 days, dense, initially white, becoming pale yellow, the centre of aerial mycelium cottony, periphery with scarce aerial mycelium, aerial mycelium formed a protuberance at center of colony.
Habitat and distribution.
Isolated from leaves of Indocalamus longiauritus in China.
Additional specimens examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Indocalamus longiauritus. 16 April 2019, S.T. Huang, HSAUP1012, HSAUP1015, and HSAUP1018 paratype; living culture SAUCC1012, SAUCC1015, and SAUCC1018.
Notes.
Strains SAUCC1011, SAUCC1012, SAUCC1015, and SAUCC1018 belong to a single species as they have similar morphological features including culture characteristics, sporodochium, and conidia, the nearly identical sequence data, and cluster in a separate branch with a good support (ML-BS: 100% and BYPP: 1.00). The species is most phylogenetically close to M. bolleyi, and their branch lengths are slightly different. Nevertheless, the morphology of M. yunnanense and M. bolleyi (Hong et al. 2008) are different in having sporodochium-like conidiomata, conidiogenous cells, and conidia. Microdochium yunnanense produced some sporodochium-like conidiomata, slimy, hyaline or orange, semi-submerged on the agar surface, with most conidial droplets formed on it and a few formed laterally along with hyaline hyphal cells. However, the conidia of M. bolleyi only formed laterally along with hyaline hyphal cells. They all produced two types of conidiogenous cells, cylindrical and ampulliform, but the size of M. yunnanense (10.0–11.5 × 1.0–2.0 μm (average 10.7 × 1.6 µm) and 6.5–10.0 × 2.5–3.4 μm (average 8.4 × 2.9 µm)), M. bolleyi (1.5–2.7 × 0.8–1.4 µm (average 2.1 × 1.0 µm) and 3.1–6.4 × 2.5–3.8 µm (average 4.9 × 3.2 µm)) were clearly different. Conidial shape was differed little, but the conidial size of M. yunnanense (6.8–10.0 × 2.4–3.5μm (average 8.3 × 3.1 µm)) has much larger than M. bolleyi (5.0–8.7 × 1.6–2.3 µm (average 6.4 × 1.9 µm)).
Microdochium
sp. indet.
3673BE11-371A-55F2-A66B-5B44B06C2F4D
Figure 4.
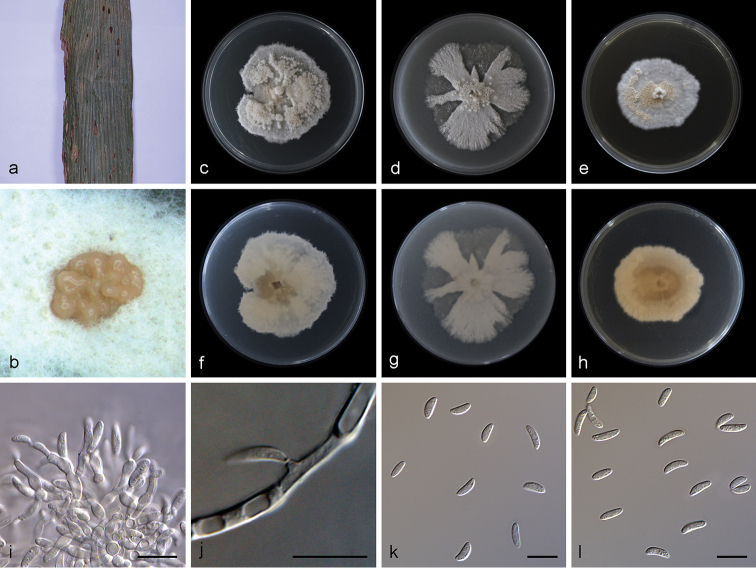
Microdochium sp. (SAUCC1017) a leaves of host plant b colony overview c–e surface of colony after 15 days on PDA (c) OA (d) MEA (e) f–h reverse of colony after 15 days on PDA (f) OA (g) MEA (h) i–k chlamydospores. Scale bars: 10μm (i–k).
Description.
Colonies on PDA attaining 73.9–80.4 mm in diameter after 15 days, felty to cottony, flat, margin entire or dentate, white, aerial mycelium abundant. Mycelium superficial, hyphae hyaline, septate, branched, smooth-walled. Reverse white to pale yellow, with yellow pigment produced with aging. Aerial hyphae aggregated to form numerous chlamydospores on agar surface. Chlamydospores thick-walled, terminal or intercalary, more frequently arranged in chains than clusters. Conidiophores not observed. Colonies on OA attaining 79.9–81.7 mm in diameter after 15 days, fluffy, margin entire, white. Reverse white. Colonies on MEA attaining 73.2–78.4 mm in diameter after 15 days, flat, with pale pink inconspicuous concentric circle near the centre, margin entire and white, aerial mycelium abundant.
Material examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Indocalamus longiauritus. 16 April 2019, S.T. Huang, HSAUP1017, living culture SAUCC1017.
Note.
Strain SAUCC1017 failed to produce conidia and lacks a complete morphological description. It formed a conspicuous independent lineage from other Microdochium species in the tree. ITS sequence BLASTn search of SAUCC1017 showed many different species with 97% identity. BLASTn searches with LSU (GenBank MH869857) sequences result in 99% identity with M. bolleyi (CBS 172.63) and TUB2 (GenBank AB625368) sequences result in 99% identity with Xylaria cubensis (strain BCC 18758). Thus, here we listed it as an unidentified species.
Discussion
Previous studies placed Microdochium in Amphisphaeriaceae (Parkinson et al. 1981; Samuels and Hallet 1983; von Arx 1984; Jaklitsch and Voglmayr 2012), which is a large heterogeneous family possessing pestalotiopsis-like asexual morphs characterised by holoblastic conidiogenous cells that produce septate, brown or hyaline conidia with appendages at both ends (Tanaka et al. 2011; Maharachchikumbura et al. 2014). Nevertheless, based on the results of phylogenetic analyses, Microdochium, Idriella, and Selenodriella were incorporated to a new family introduced as Microdochiaceae by Hernández-Restrepo et al. (2016), which is characterised by asexual morphs that produce polyblastic, sympodial or annellidic conidiogenous cells with hyaline conidia without appendages and sexual morphs that are monographella-like. In Microdochium, the color of conidiogenous cells is hyaline, and the shape of conidia seem to be taxonomic important feature. The conidial shape of Microdochium is more variable from cylindrical, fusoid or oblong, to lunate, straight or curved, with truncate bases and apices mainly rounded.
Three sections were widely accepted in Microdochium based on the type of conidiogenous cells and conidia by Braun (1995) and Hernández-Restrepo et al. (2016). Type I: Microdochium sect. Gerlachia forming annellidic conidiogenous cells with percurrent proliferations; Type II: Microdochium forming sympodial, often subdenticulate conidiogenous cells, and relatively more or less fusiform, straight to somewhat curved or falcate, 0–3-septate or even pluriseptate conidia; and Type III: Gloeocercospora forming sympodial conidiogenous cells, and very long, scolecosporous and pluriseptate conidia.
From the previous molecular studies of Microdochium (Jaklitsch and Voglmayr 2012; Jewell and Hsiang 2013; Zhang et al. 2015), the four gene regions (ITS, LSU, RPB2, TUB2) were chosen in this study. The LSU is informative enough for generic placement of Microdochium. The individual gene regions of ITS, TUB2, and RPB2 proved to be able to resolve species in Microdochium (results not shown). However, TUB2 was the more informative than other gene regions and showed longer distances between species and higher support values. This results in our study agree with previous studies in other xylariaceous genera (Hsieh et al. 2005; Læssøe et al. 2013; Hernández-Restrepo et al. 2016). By combining phylogenetic analysis and morphology, two species of Microdochium were delimited as new species, namely M. yunnanense sp. nov. and M. indocalami sp. nov. In order to support the validity of these new species, we followed the guidelines of Hernández-Restrepo et al. (2016).
Supplementary Material
Acknowledgements
This work was jointly supported by the National Natural Science Foundation of China (no. 31770016, 31750001, and 31900014) and the China Postdoctoral Science Foundation (no. 2018M632699).
Citation
Huang S, Xia J, Zhang X, Sun W, Li Z (2020) Two new species of Microdochium from Indocalamus longiauritus in south-western China. MycoKeys 72: 93–108. https://doi.org/10.3897/mycokeys.72.55445
Contributor Information
Wenxiu Sun, Email: wxsun@yangtzeu.edu.cn.
Zhuang Li, Email: junwuxue@126.com.
References
- von Arx JA. (1984) Notes on Monographella and Microdochium. Transactions of The British Mycological Society 83(2): 373–374. 10.1016/S0007-1536(84)80168-0 [DOI] [Google Scholar]
- Braun U. (1995) A Monograph of Cercosporella, Ramularia and allied genera (phytopathogenic Hyphomycetes) (Vol. 1). IHW-Verlag, Eching, 494 pp. [Google Scholar]
- Cai L, Hyde KD, Taylor PWJ, Weir B, Waller J, Abang MM, Zhang ZJ, Yang YL, Phoulivong S, Liu ZY, Prihastuti H, Shivas RG, McKenzie EHC, Johnston PR. (2009) A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 183–204. [Google Scholar]
- Choi YW, Hyde KD, WH Ho. (1999) Single spore isolation of fungi. Fungal Diversity 3: 29–38. [Google Scholar]
- Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu JC, Liu XZ, Stadler M. (2014) The sooty moulds. Fungal Diversity 66(1): 1–36. 10.1007/s13225-014-0278-5 [DOI] [Google Scholar]
- Crous PW, Schumacher RK, Akulov A, Thangavel R, Hernández-Restrepo M, Carnegie AJ, Cheewangkoon R, Wingfield MJ, Summerell BA, Quaedvlieg W. (2019) New and interesting fungi 2. Fungal Systematics and Evolution 3(1): 57–134. 10.3114/fuse.2019.03.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, Akulov A, Denman S, Roux J, Braun U, Burgess TI, Carnegie AJ, Váczy KZ. (2018) New and interesting fungi 1. Fungal Systematics and Evolution 1(1): 169–215. 10.3114/fuse.2018.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YH, Sun W, Su YY, Cai L. (2014) Three new species of Phomopsis in Gutianshan Nature Reserve in China. Mycological Progress 13(1): 111–121. 10.1007/s11557-013-0898-2 [DOI] [Google Scholar]
- Glynn NC, Hare MC, Parry DW, Edwards SG. (2005) Phylogenetic analysis of EF-1 alpha gene sequences from isolates of Microdochium Nivale leads to elevation of varieties Majus and Nivale to species status. Fungal Biology 109(8): 872–880. 10.1017/S0953756205003370 [DOI] [PubMed] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. (2000) Fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytologist 147(3): 617–630. 10.1046/j.1469-8137.2000.00716.x [DOI] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Groenewald JZ, Crous PW. (2016) Taxonomic and phylogenetic re-evaluation of Microdochium, Monographella and Idriella. Persoonia 36(1): 57–82. 10.3767/003158516X688676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SK, Kim WG, Choi HW, Lee SY. (2008) Identification of Microdochium bolleyi associated with basal rot of creeping bent grass in Korea. Mycobiology 36(2): 77–80. 10.4489/MYCO.2008.36.2.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh HM, Ju YM, Rogers JD. (2005) Molecular phylogeny of Hypoxylon and closely related genera. Mycologia 97(4): 844–865. 10.1080/15572536.2006.11832776 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17(17): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Voglmayr H. (2012) Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Diversity 52(1): 75–98. 10.1007/s13225-011-0104-2 [DOI] [Google Scholar]
- Jewell LE, Hsiang T. (2013) Multigene differences between Microdochium Nivale and Microdochium majus. Botany 91(2): 99–106. 10.1139/cjb-2012-0178 [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasna H, Bateman GL. (2007) Microdochium Triticicola sp. nov. from roots of Triticum aestivum in the United Kingdom. Mycologia 99(5): 765–776. 10.3852/mycologia.99.5.765 [DOI] [PubMed] [Google Scholar]
- Læssøe T, Srikitikulchai P, Luangsa-ard JJD, Stadler M. (2013) Theissenia reconsidered, including molecular phylogeny of the type species T. Pyrenocrata and a new genus Durotheca (Xylariaceae, Ascomycota). IMA Fungus 4(1): 57–69. 10.5598/imafungus.2013.04.01.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FF, Weir BS, Damm U, Crous PW, Wang Y, Liu B, Wang M, Zhang MM, Cai L. (2015) Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. Gloeosporioides complex. Persoonia 35: 63–86. 10.3767/003158515X687597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic Relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16(12): 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Maharachchikumbura SSN, Hyde KD, Groenewald JZ. (2014) Pestalotiopsis revisited. Studies in Mycology 79: 121–186. 10.1016/j.simyco.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Felix Y, Hernández-Restrepo M, Wingfield MJ, Akulov A, Carnegie AJ, Cheewangkoon R, Gramaje D, Groenewald JZ, Guarnaccia V, Halleen F. (2019) Genera of phytopathogenic fungi: GOPHY 2. Studies in Mycology 92: 47–133. 10.1016/j.simyco.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment. Bridging from the extreme to the campus and beyond. Association for Computing Machinery, USA, 8 pp 10.1145/2335755.2335836 [DOI] [Google Scholar]
- Nylander JAA. (2004) MrModelTest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Parkinson VO, Sivanesan A, Booth C. (1981) The perfect state of the rice leaf scald fungus and the taxonomy of both the perfect and imperfect states. Transactions of The British Mycological Society 76(1): 59–69. 10.1016/S0007-1536(81)80009-5 [DOI] [Google Scholar]
- Petrak F. (1924) Mykologische Notizen. VII (No. 144–145). Annales Mycologici 22(1–2): 1–182. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels GJ, Hallett IC. (1983) Microdochium stoveri and Monographella stoveri, new combinations for Fusarium stoveri and Micronectriella stoveri. Transactions of The British Mycological Society 81(3): 473–483. 10.1016/S0007-1536(83)80115-6 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung GH, Sung JM, Hywel-Jones NL, Spatafora JW. (2007) A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution 44(3): 1204–1223. 10.1016/j.ympev.2007.03.011 [DOI] [PubMed] [Google Scholar]
- Sydow H. (1924) Mycotheca germanic. Fasc. XLII–XLV (No. 2051–2250). Annales Mycologici 22: 257–268. [Google Scholar]
- Tanaka K, Endo M, Hirayama K, Okane I, Hosoya T, Sato T. (2011) Phylogeny of Discosia and Seimatosporium, and introduction of Adisciso and Immersidiscosia genera nova. Persoonia 26(1): 85–98. 10.3767/003158511X576666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor FJRM, Lee SH, Taylor L, Shawe-Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. Innis MA, Gelfand DH, Sninsky JJ (Eds) PCR Protocols: a Guide to Methods and Applications. Academic Press Inc., New York, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Zhang K, Su YY, Cai L. (2013) An optimized protocol of single spore isolation for fungi. Cryptogamie Mycologie 34(4): 349–356. 10.7872/crym.v34.iss4.2013.349 [DOI] [Google Scholar]
- Zhang W, Nan ZB, Tian P, Hu MJ, Gao ZY, Li M, Liu GD. (2015) Microdochium Paspali, a new species causing seashore paspalum disease in southern China. Mycologia 107(1): 80–89. 10.3852/14-119 [DOI] [PubMed] [Google Scholar]
- Zhang ZF, Liu F, Zhou X, Liu XZ, Liu SJ, Cai L. (2017) Culturable Mycobiota from karst caves in China, with descriptions of 20 new species. Persoonia 39(1): 1–31. 10.3767/persoonia.2017.39.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.