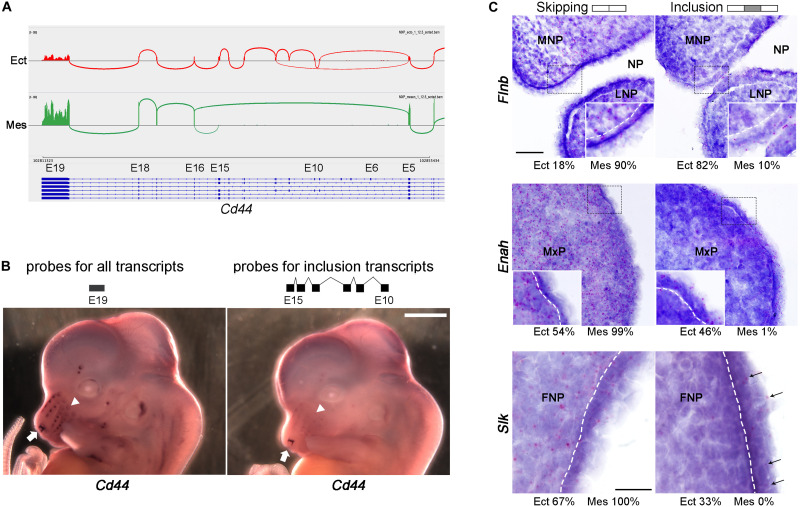
FIGURE 4.
In situ hybridization validation of SE events across tissue layers. (A) Sashimi plot showing skipping of multiple exons (exons 6–15) of Cd44 in mesenchyme. Sashimi plot was generated by using IGV browser with RNA-seq.bam files of E12.5 MxP ectoderm (Ect) sample 1 (top track, red) and E12.5 MxP mesenchyme (Mes) sample 1 (bottom track, green). The vertical piles are RNA-seq reads of exons, while the arcs connecting exons show how the exons are spliced. The isoform models are shown under the tracks with blue rectangles as exons, lines as introns, and arrowheads indicating the direction of transcripts (right to left). Exon (E) numbers of Cd44 transcript variant 1 (NM_009851.2) are marked on top of the transcripts. Note that only the splicing junctions with five or more reads are shown here. (B) Whole mount in situ hybridization of E12.5 embryos for an exon 19 probe detecting all Cd44 transcripts (left panel) or an exons 10–15 probe specific for ectodermal transcripts (right panel). The arrows point to the nasal cavity and the arrowheads point to vibrissae. Scale bar: 1 mm. (C) In situ hybridization on E11.5 frontal sections of FNP, MxP, and FNP using Basescope for SE events in Flnb, Enah, and Slk, respectively. Gene names are shown on the left and probes (skipping or inclusion) on top. The rMATS estimated average percentage of skipping and inclusion for Ect and Mes tissues (FNP for Flnb, MxP for Enah, and FNP for Slk) are shown under the corresponding panels. Red punctuate dots are signals for the Basescope probes. The sections are counterstained with hematoxylin. For all three genes the isoforms with the skipped exon are expressed in both ectoderm and mesenchyme, while the isoforms which include the alternative exon are mainly expressed in the ectoderm. Insets in Flnb and Enah panels show enlarged images of the areas in the dashed lined rectangles. White dashed lines indicate the boundary between ectoderm and mesenchyme. In the Slk panels, arrows indicate Basescope signals for the probe detecting the alternative exon. Scale bar is 50 μm for the Flnb and Enah panels, and scale bar for Slk is 20 μm. FNP, frontonasal process; LNP, lateral nasal process; MNP, medial nasal process; MxP, maxillary process; NP, nasal pit.