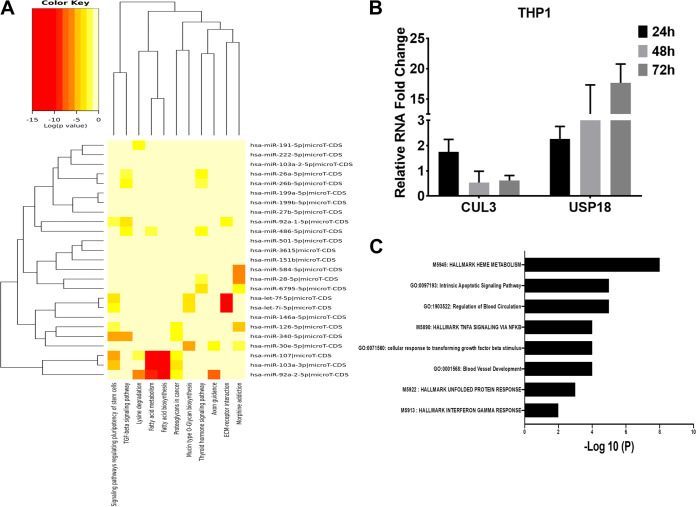
FIG 7.
Dysregulated miRNAs affecting the global metabolic pathways and validation of target genes. The miRNA target gene and pathway enrichment analyses were performed using miRPath v.3. (A) Heatmap represents miRNAs and targeted pathways. Clustering is based on significance levels. Darker colors represent a higher significance. On the vertical axis, miRNAs displaying similar pathway targeting patterns are clustered together. (B) Determining the expression of CUL3 and USP18 in DENV-infected cell lines. THP1 cells, infected at a multiplicity of infection (MOI) of 3, were harvested at 24 h, 48 h, and 72 h p.i. The total RNA from cells was isolated, and relative levels of viral RNA, CUL3, and USP18 genes were determined by qPCR. (C) Pathway enrichment analysis of genes dysregulated in PBMCs of dengue-infected patients and targeted by the miRNAs. Pathways are plotted against the P values.