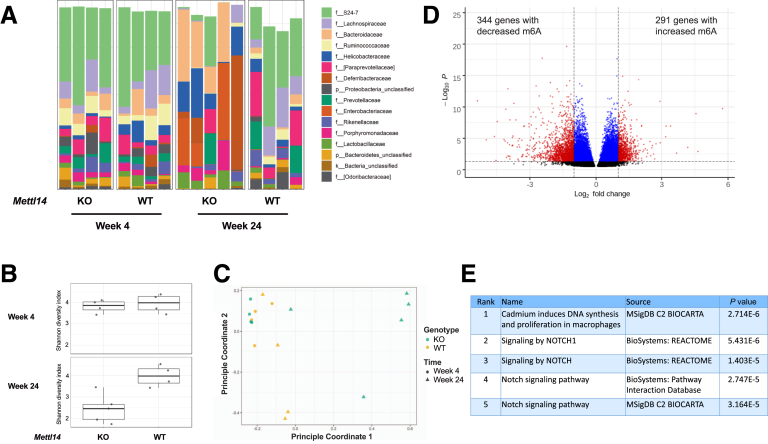
Figure 8.
Microbiome analysis of the CD4-Cre+/TgMettl14FL/FL conditional knockout mice. (A) Taxonomic composition, (B) alpha diversity, and (C) principal coordinate analysis plot of beta diversity of CD4-Cre+/TgMettl14FL/FL conditional knockout mice compared with control animals at week 4 and week 24. WT indicates wild-type littermate control. KO indicates CD4-Cre+/TgMettl14FL/FL conditional knockout. n = 4–5 mice per group. (D) m6A profiling of CD4+ T cells from germ-free mice compared with CD4+ T cells from specific-pathogen-free mice. Black color indicates nonsignificant; blue color indicates P < .05 and absolute Log2 fold change < 1; red color indicates P < .05 and absolute Log2 fold change > 1. (E) Significant pathways represented by genes with differential m6A levels. The top 5 most significant pathways were shown.