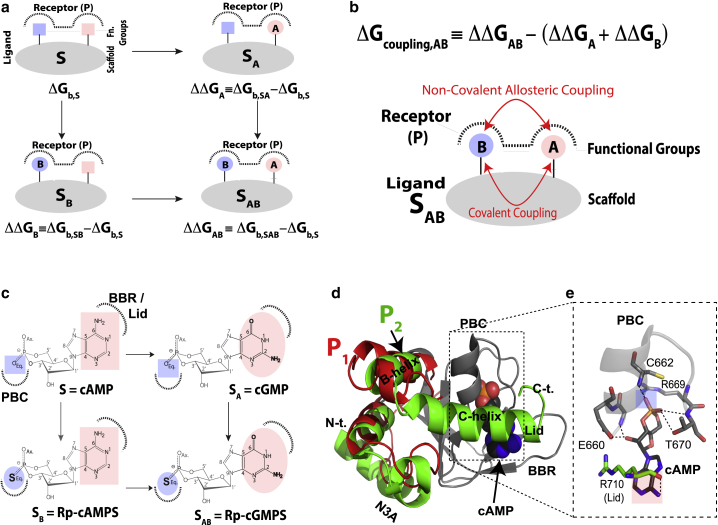
Figure 1.
Nonadditivity between two ligand substituents as defined by double-ligand cycles. (a) General double-ligand cycle for a ligand composed of a scaffold or linker S (gray) and two substituents A (pink) and B (blue). The cycle includes four protein complexes in which the receptor (P) binds either S, the single-substituted ligands (i.e. SA or SB), or the double-substituted ligand SAB. Dotted lines denote binding sites in P. ΔGb,S is the binding free energy for ligand S, whereas ΔΔGA is defined as the contribution to binding arising from substituent A in ligand SA. Similar definitions apply to SB and SAB. If modifications A and B are independent, then ΔΔGAB = ΔΔGA + ΔΔGB, which is the equation defining binding additivity (Fig. S1 a). (b) Definition of the coupling free energy between substituents A and B (ΔGcoupling,AB) and a scheme illustrating the covalent and/or noncovalent double nature of such coupling. (c) Example of the double-ligand cycle including the cyclic nucleotide monophosphates (cNMPs) cAMP, the phosphorothioate Rp-cAMPS, cGMP, and Rp-cGMPS. (d) Autoinhibitory and active conformations accessed by the cNMP-binding domain (CNBD) of HCN4 in the apo and cAMP-bound forms (PDB: 2MNG and PDB: 3OTF; denoted here as P1 and P2, respectively). The invariant β-subdomain is shown as a gray cartoon, whereas the more dynamic α-subdomain is depicted in red (apo) or green (cAMP-bound). Key structural motifs are labeled, including the phosphate-binding cassette (PBC), the base-binding region (BBR), and the N3A. (e) Expanded view of the PBC and part of the lid region of the HCN4 CNBD, highlighting key interactions of these regions with cAMP. Hydrogen bonds are shown as dashed lines. To see this figure in color, go online.