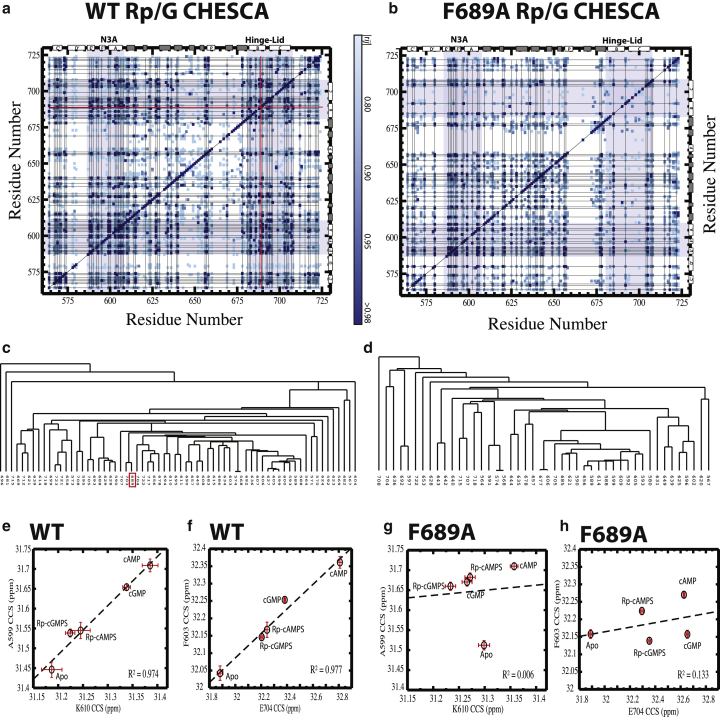
Figure 4.
Chemical shift covariance analysis (CHESCA) of the HCN4 CNBD based on the Rp/G double-ligand cycle. (a) CHESCA correlation matrix for wt HCN4 (563–724) generated using the apo sample and the four cNMPs in the Rp/G cycle of Fig. 1c. The secondary structure is shown as white (α-helices) and gray (β-strands) rectangles. The grid lines depict the single-linkage allosteric cluster of (c), whereas the red lines mark the position of F689. Shaded areas mark the N3A and hinge-lid helical regions. (b) As (a) but for the F689A HCN4 (563–724) mutant. (c) Dendrogram of the cluster obtained through single-linkage agglomerative clustering for the wt HCN4 CNBD. A Pearson’s correlation coefficient cutoff ≥0.98 was utilized. F689 is highlighted by a red box. (d) As (c) but for the F689A HCN4 (563–724) mutant. (e and f) Representative pairwise interresidue combined chemical shift correlations for wt HCN4 (563–724). (g and h) Pairwise plots for the same residue pairs in (e) and (f) are shown but for F689A HCN4 (563–724), illustrating the decorrelation caused by the mutation, which perturbs the allosteric networks of wt HCN4. To see this figure in color, go online. Errors in the chemical shifts were assessed as previously described (40,41).