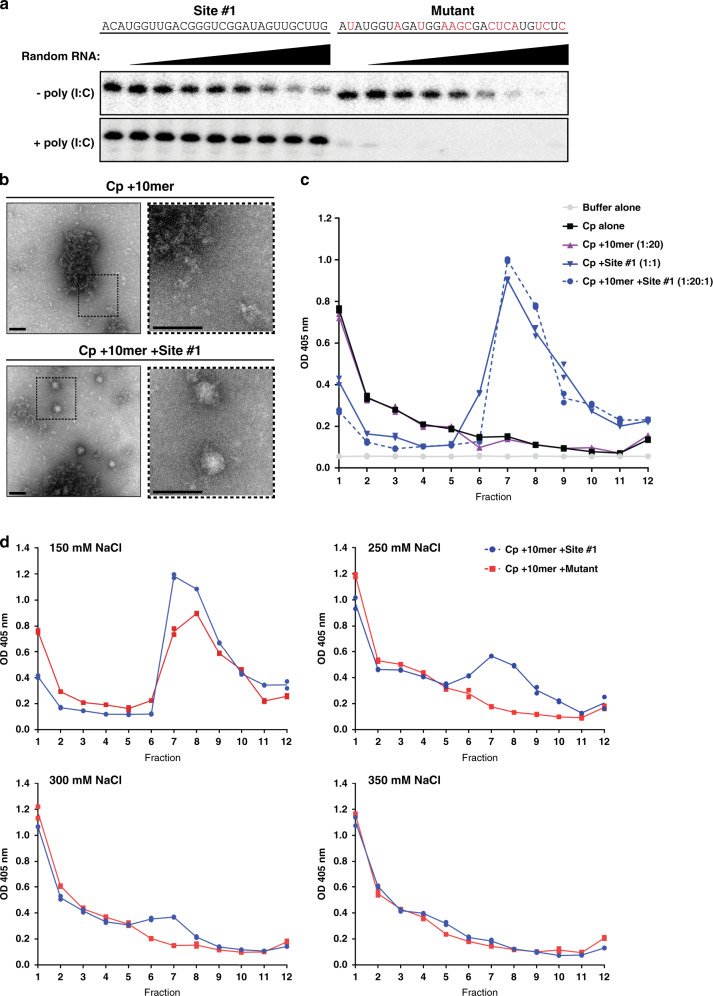
Fig. 6. Cp specifically binds and assembles with site #1 in vitro.
a Upper panel: 333 nM 32P-labeled site #1 (left) or mutant (right; mutated residues highlighted in red) RNAs were bound to 13.3 pmole Cp immobilized on Streptactin beads in a 30-µL reaction. Increasing concentrations (4–1000 nM) of random RNA were added as a competitor. Bound RNAs were extracted and subjected to Urea-PAGE followed by autoradiography. Lower panel: as above, but immobilized Cp was first preincubated with 20 μg/mL of poly(I:C) to shield non-specific electrostatic interactions. Results are representative of two independent experiments. b Negative stain EM of CLP assembly reactions of Cp +10mer or Cp +10mer +Site #1 RNA. Dashed boxes in the right panel are ×3 magnification of the corresponding left panel. Scale bar is 80 nm. Images represent one experiment. c CLP assembly reactions were incubated for 30 min at 25 °C (150 mM NaCl) and then analyzed by sucrose gradient sedimentation. Cp from aliquots of fractions was detected by ELISA. Individual data points from technical duplicates are plotted and the line represents their mean. Fraction 1 is the top of the gradient. Results are representative of n = 4 independent experiments for Cp alone, Cp +10mer (1:20), Cp +Site #1 (1:1), and Cp +10mer +Site #1 (1:20:1), and n = 2 independent experiments for Buffer alone. d As in c, except the indicated salt concentrations were used in CLP assembly reactions comparing CLP assembly with site #1 or mutant RNAs. Individual data points from technical duplicates are plotted and the line represents their mean. Results are representative of n = 4 independent experiments for 250 mM NaCl, and n = 2 independent experiments for 150, 300, and 350 mM NaCl. See also Supplementary Fig. 5.