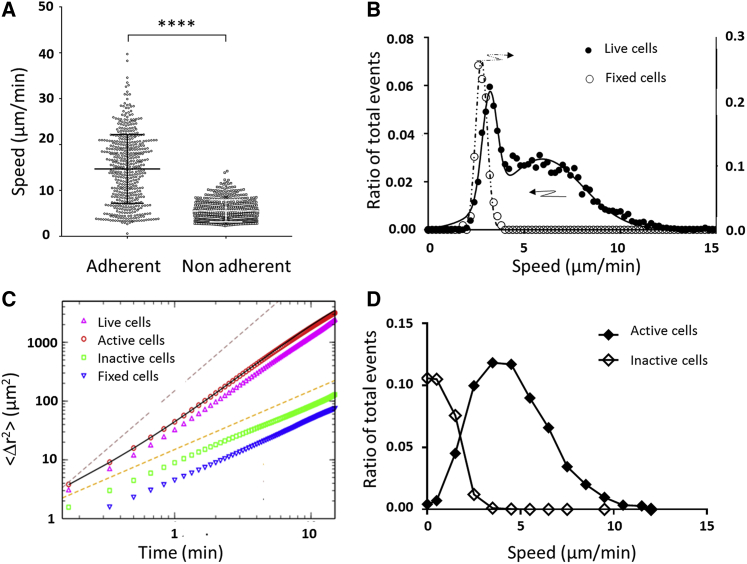
Figure 2.
Active polarized cells migrate without adhesion in 2D at 5 μm min−1. (A) Raw curvilinear speed of cells crawling on adherent ICAM-1-treated substrates and migrating on antiadherent Pluronic F127-treated substrates, estimated by averaging the displacements of cell mass centers over intervals of 30 s. Bars are standard deviations. N = 500 cells, ****p < 0.0001 (two-tailed Student’s t-test). (B) Histogram of raw curvilinear speed of live nonadherent migrating cells (filled dots, left y axis) and fixed cells (hollow dots, right y axis). Data are fitted with a single Gaussian for fixed cells (dotted line) and a double Gaussian for live cells (dark line). Live cells are composed of one population of diffusing cells and one of migrating cells with an average speed of 5.9 μm min−1. (C) Mean-square displacement <[r(t) − r(0)]2> as a function of time for all cells and all steps was combined in the case of live cells (upward pointed triangles) and fixed cells (downward pointed triangles). Fixed cells have purely diffusive behavior corresponding to Dt = 2.34 μm2 min−1. Circles and squares show the mean-square displacement for active and inactive cells, respectively. Black line is a fit of active cells using Eq. 9 (Supporting Materials and Methods) with vs = 4.3 μm min−1, Dr = 0.19 min−1, and Dt = 7.28 μm2 min−1. Dashed lines show the limit cases of purely ballistic and diffusive behavior with slopes of, respectively, 2 and 1. (D) Histogram of average speed per cell determined using Eq. 1 as in (C) for active cells (filled diamonds) and inactive cells (hollow diamonds). Root mean-square speed of active cells is equal to 4.9 μm min−1. Lines are guides for the eye. To see this figure in color, go online.