Figure 4.
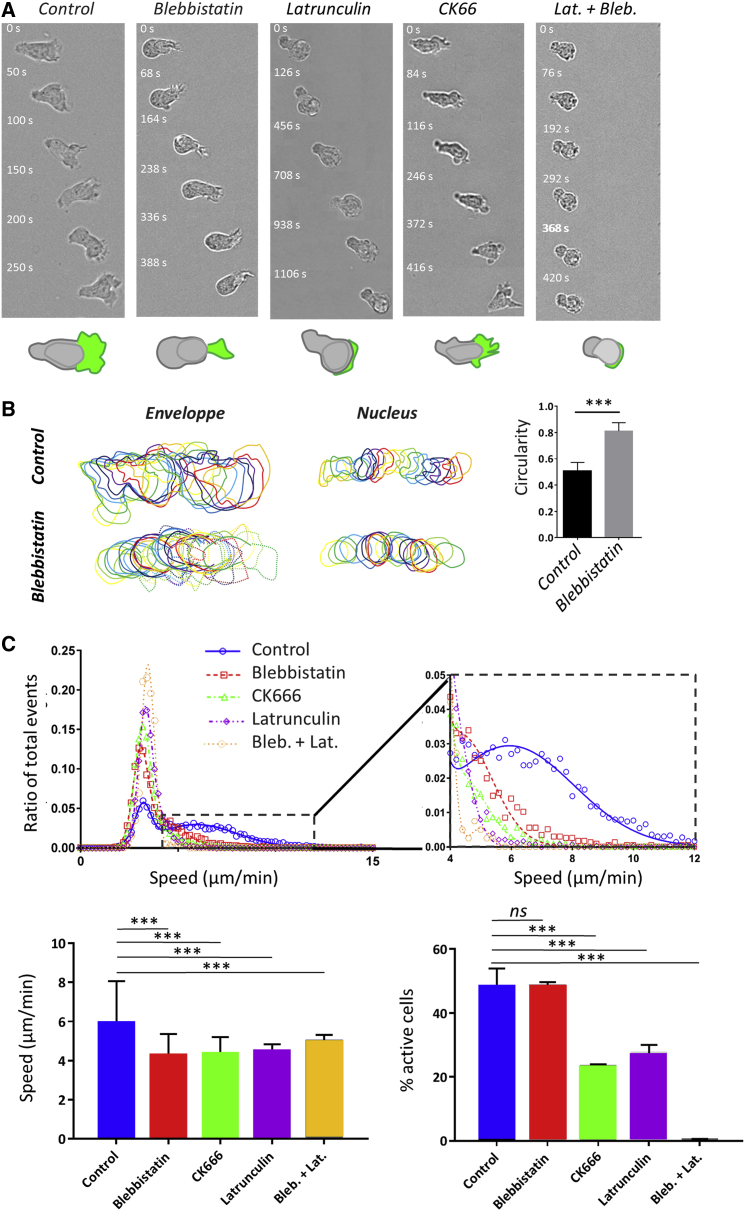
Actin propels swimming more by polymerization than by contractility. (A) Bright-field image sequences show the shape and dynamics of cells swimming on an antiadhesive substrate versus the addition of actomyosin inhibitors. Shown from left to right: wild-type cells and cells treated with 50 μM blebbistatin, 0.05 μM latrunculin, 100 μM CK666, and combined latrunculin and blebbistatin. See also Video S6. Cartoons at the bottom reproduce the cell in the first image to illustrate in each case the shape of the cell body (rear and nucleus) in gray and of the cell front or lamellipod in green. Blebbistatin-treated cells have a roundish cell body without traveling protrusion and a reduced but active lamellipod; latrunculin-treated cells have almost no lamellipod; and CK666-treated cells have a perturbed lamellipod forming blebs and spikes. Cells treated with blebbistatin and latruculin have a roundish noncontractile cell body and no lamellipod (Scale bars, 10 μm). (B) Left: shown are representative sequences of envelope and nucleus contours of a control cell (top) and of a cell treated with 50 μM blebbistatin (bottom), suggesting a difference of contractile activity. Time lag between each contour is 10 s. Right: shown is the quantification of the circularity of cells envelops for control cells and 50 μM blebbistatin treatment on image sequence taken at magnification ×60 and cell contour determined using Ilastik. Error bars correspond to standard deviation. Cells N >10, and experiments N = 3. ∗∗∗p < 0.001 (two-tailed Student’s t-test). (C) Top: shown is a histogram of raw curvilinear speeds for swimming cells in response to above-mentioned actin inhibitors treatments. The lines correspond to a fit by a double Gaussian function. Insert presents a zoom of the histogram for the active cells corresponding to the Gaussian of high mean speed. Bottom left: shown is the average speed and SD corresponding to the Gaussian of high mean speed. Bottom right: shown is the percentage of active motile cells, and error bars correspond to SEM. Cells N = 4342 (HBSS), 2353 (blebbistatin), 5582 (CK666), 2255 (latrunculin), and 403 (blebbistatin plus latrunculin). Error bars correspond to SEM. Experiments N >5. ∗∗∗p < 0.001 with respect to homogeneous substrate, one-way ANOVA followed by the Tukey multiple comparison test. To see this figure in color, go online.