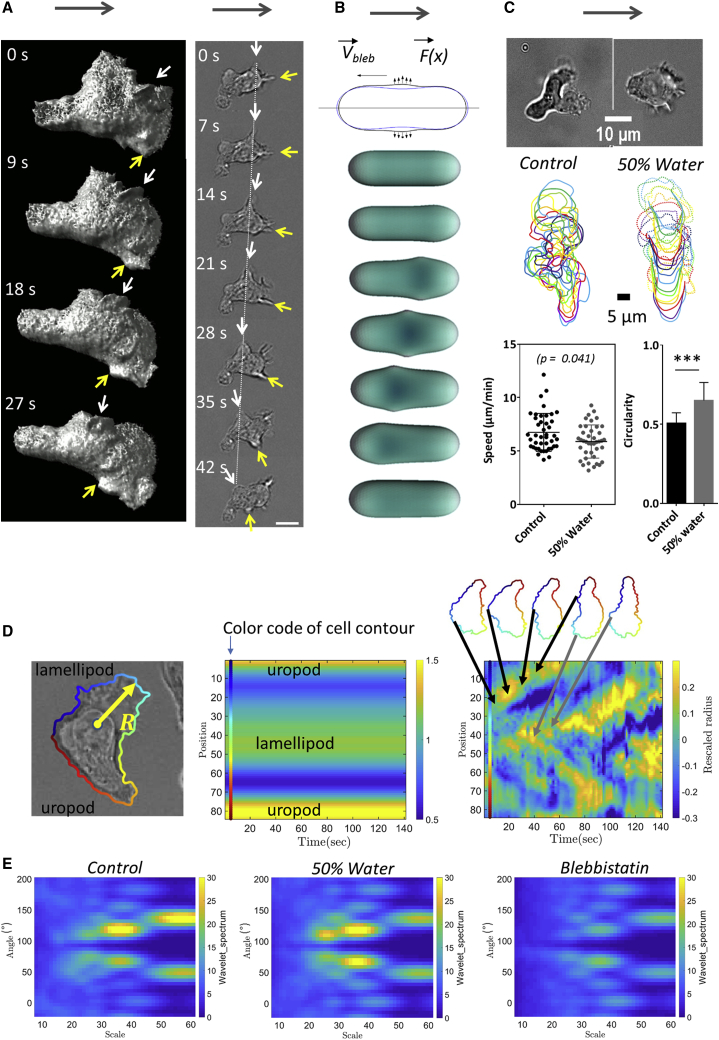
Figure 5.
Protrusion paddling alone seems not efficient enough to propel swimming. (A) Image sequence of swimming cell with micron-scale protrusions traveling along cell body. Left: soSPIM images of a cell transfected with RFP-Lifeact reveals the shape and motion of waves of actin protrusion in three dimensions. White and yellow arrows point to particular protrusions. See also Video S7. Right: bright-field images of a swimming cell show dynamics of protrusions. See also Video S8. Protrusions (white and yellow arrows) travel backward in the frame of the cell and of the lab (white dashed line). Gray arrows indicate swimming direction. Scale bars, 10 μm. (B) Schematic illustrates the model of cell swimming by protrusive blebs. Top: blue and black contours are the initial and deformed configurations of the cell in the model. Bottom: shown is the sequence of cell shapes obtained by the numerical simulation. Simulations yield that a cell propelled by shape waves is 1000 times slower than the protrusion wave (details in Supporting Materials and Methods). (C) Osmotic swelling affects cell shape but not swimming speed. Top: representative images of a cell in medium and in water dilution at 50%. Middle: representative cell contour sequences (time lag is 10 s between each contour). The cell swollen by osmotic stress displays less deformations of cell body (full lines), but lamellipod protrusions are still distinguishable (dotted line). Bottom: shown is the swimming speed in medium and with water dilution at 50%. (cells N >30, p value of t-test) and circularity of cells on image taken at magnification ×60, with cell contours determined using Ilastik. Error bars stand for standard deviation Cell N >10, and experiments N = 3. ∗∗∗p < 0.001 (two-tailed Student’s t-test). See also Video S10. (D) Measurement of cell protrusion dynamics. Left: Shown is a representative example of an elliptic Fourier contour reconstruction. Radius R definition of edge distance to the center of mass is shown. Middle and right: shown are kymographs of rescaled R versus position along the cell perimeter, either averaged in time (middle) or with time-average contribution subtracted (right). Color scale on the left corresponds to the color code used in contour reconstruction, allowing one to identify the position in kymographs of lamellipod, uropod, and propagating protrusions. See also Fig. S8. (E) Spatiotemporal spectrum of protrusion dynamics by wavelet transform. Shown is the average wavelet spectra of rescaled R kymographs without time-averaged contribution (see also Fig. S9). To see this figure in color, go online.