Summary
Prospective molecular characterization of cancer has enabled physicians to define the genomic changes of each patient’s tumor in real-time and select personalized therapies based on these detailed portraits. Despite the promise of such an approach, previously unrecognized biologic and therapeutic complexity is emerging. Here, we synthesize lessons learned and discuss the steps required to extend the benefits of genome-driven oncology, including proposing strategies for improved drug design, more nuanced patient selection, and optimized use of available therapies. Finally, we suggest ways that next-generation genome-driven clinical trials can evolve to accelerate our understanding of cancer biology and improve patient outcomes.
Keywords: precision oncology, drug development, clinical trial design, genome-driven oncology
Introduction
The scope of precision oncology is rapidly expanding to address previously undruggable targets and rare genomic drivers (Hyman et al., 2017b). While only a minority of patients currently benefit from genomic matching to targeted therapies, this population continues to grow as the field advances (Hyman et al., 2017b; Schram and Hyman, 2017; Zehir et al., 2017). Herein, we propose next steps for refining drug development, patient matching to molecularly-guided therapies, and clinical trial design to extend the benefits of precision oncology. We focus in particular on genomically-driven drug development, which complements research on immunotherapy and other novel treatment strategies.
Developing Better Drugs
The current generation of therapies that define precision oncology have helped to elucidate many of the key characteristics of an optimal molecularly-targeted drug. Among the most important factors are its therapeutic index, target selectivity, and resistance liabilities.
Therapeutic Index
The presence of a therapeutic window to allow for optimal dosing is crucial for the success of a targeted therapy. The therapeutic index is a function of drug selectivity, characteristics of the target, and off-target toxicities. For example, the therapeutic index of EGFR inhibitors varies based on differences in selectivity between the activating mutation being targeted and wildtype EGFR. Many patients that respond well to first- and second-generation EGFR inhibitors such as erlotinib, gefitinib, and afatinib, have L858R mutations and exon 19 deletions that increase receptor dimerization and diminish ATP binding, enhancing the affinity of inhibitors compared to wildtype EGFR. By comparison, these agents have a negative therapeutic index in EGFR exon 20 insertions because inhibition against exon 20 mutants is less potent than against wildtype EGFR, limiting the tolerability of this class of agents (Beau-Faller et al., 2014; Naidoo et al., 2015; Robichaux et al., 2018; Vyse and Huang, 2019; Yun et al., 2007). Alternative irreversible inhibitors such as TAK-788 and poziotinib have been developed that have similar potency against wildtype and exon 20 mutant EGFR which may provide sufficient therapeutic index to permit anti-tumor activity (Chouitar et al., 2018; Han et al., 2017; Vyse and Huang, 2019). While this approach has for the first time unlocked clinical activity in previously refractory EGFR exon 20 mutant non-small cell lung cancers (NSCLCs), relatively narrow therapeutic indices manifest clinically as EGFR-mediated toxicities (Han et al., 2017). The challenge of developing drugs for patients with EGFR exon 20 mutations reveals that for certain mutations, substantial toxicity may derive from inhibition of the wildtype target in normal host tissues (so called on-target, off-tumor effects).
Beyond the importance of therapeutic index for individual molecularly targeted drugs, toxicity has also been the primary barrier to successfully implementing targeted therapy combinations. For example, despite very promising preclinical data suggesting concurrent MAPK and PI3K pathway inhibition prevents feedback reactivation and bypass resistance, clinical attempts to co-target these pathways have proven largely intolerable (Lopez and Banerji, 2017; Shimizu et al., 2012). Careful attention to both the individual therapeutic indices of drugs and the extent of overlapping toxicities is critical when choosing therapeutic combinations.
Target Selectivity
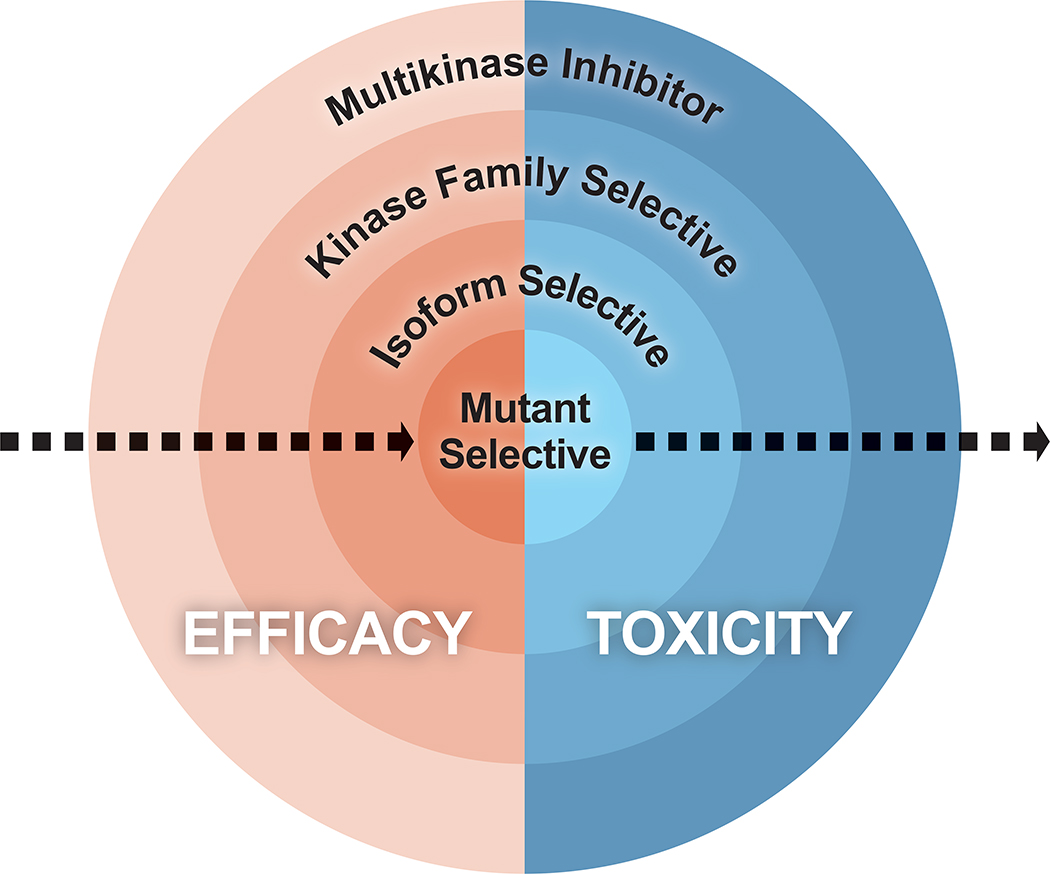
The development of purpose-built inhibitors that selectively and potently inhibit the target of interest has also unlocked therapeutic potential for a broader proportion of cancer patients. Target selectivity decreases off-target toxicity and allows for more potent drug activity, thereby improving efficacy (Figure 1). Activating RET alterations including fusions are found in ~2% of lung adenocarcinoma and up to 20% of papillary thyroid cancers, while activating germline and somatic mutations are identified in the majority of medullary thyroid carcinomas (Drilon et al., 2018a; Dvorakova et al., 2008; Elisei et al., 2008; Kato et al., 2017; Moura et al., 2009; Mulligan, 2014; Stransky et al., 2014; Subbiah et al., 2018a; Subbiah et al., 2018b). Historically, multikinase inhibitors (MKI) that include some degree of RET inhibition such as lenvatinib, vandetanib, cabozatinib, and ponatinib have exhibited limited to modest clinical activity in RET altered tumors, depending on the patient population. All of these drugs exhibit more potent off-target inhibition, typically VEGFR (KDR), that defines their dose limiting toxicity and thus precludes maximal RET blockade (Azad et al., 2009; Drilon et al., 2018a; Drilon et al., 2016; Elisei et al., 2013; Gautschi et al., 2017; Hayman et al., 2012; Mologni et al., 2013; Subbiah et al., 2018b; Wirth et al., 2018). Conversely, selective RET inhibitors, including selpercatinib (LOXO-292) and pralsetinib (BLU-667), have been developed that permit potent and tonic target inhibition (Brandhuber BB; Subbiah et al., 2018a) and have demonstrated substantial efficacy and favorable safety profiles in comparison to MKIs (Ackermann et al., 2019; Drilon et al., 2019; Drilon et al., 2016; Drilon et al., 2018c; Gainor et al., 2019; Gautschi et al., 2017; Guo et al., 2019; Subbiah et al., 2018a; Subbiah et al., 2018b; Taylor et al., 2019; Wirth et al., 2019). Ultimately, improved understanding of the genomic drivers of individual cancers coupled with advances in structural biology have enabled the development of rational, fit-for-purpose drugs that specifically target the biomarker of interest. The creation of such selective inhibitors is critical to optimize tolerability and maximize therapeutic efficacy.
Figure 1. Relationship between efficacy and toxicity of different drug classes.
In general, efficacy increases and toxicity decreases as kinase inhibitors become more specific for the mutated protein being targeted.
Resistance Liabilities
Potential mechanisms of primary and acquired resistance should be accounted for when designing drugs. Considerations include both resistance resulting from anatomic sanctuary sites with poor drug penetrance as well as resistance secondary to molecular alterations. For cancers where metastasis to the brain are common including NSCLC, breast cancer, and melanoma, ensuring that drugs targeting key genomic alterations in these cancers have adequate central nervous system (CNS) penetrance has become a key design parameter (Offin et al., 2019). While crizotinib, a first-generation ALK inhibitor, achieves a high initial rate of systemic disease control, poor brain penetration leads to as many as 60% of patients developing CNS progression while on therapy (Johung et al., 2016; Zhang et al., 2015; Zweig and Neal, 2018). Prospective evaluation of CNS penetrant next-generation ALK inhibitors have demonstrated markedly improved disease control within the brain, ultimately contributing to increased progression-free and overall survival (Camidge et al., 2018; Gadgeel et al., 2018; Peters et al., 2017; Shaw et al., 2016).
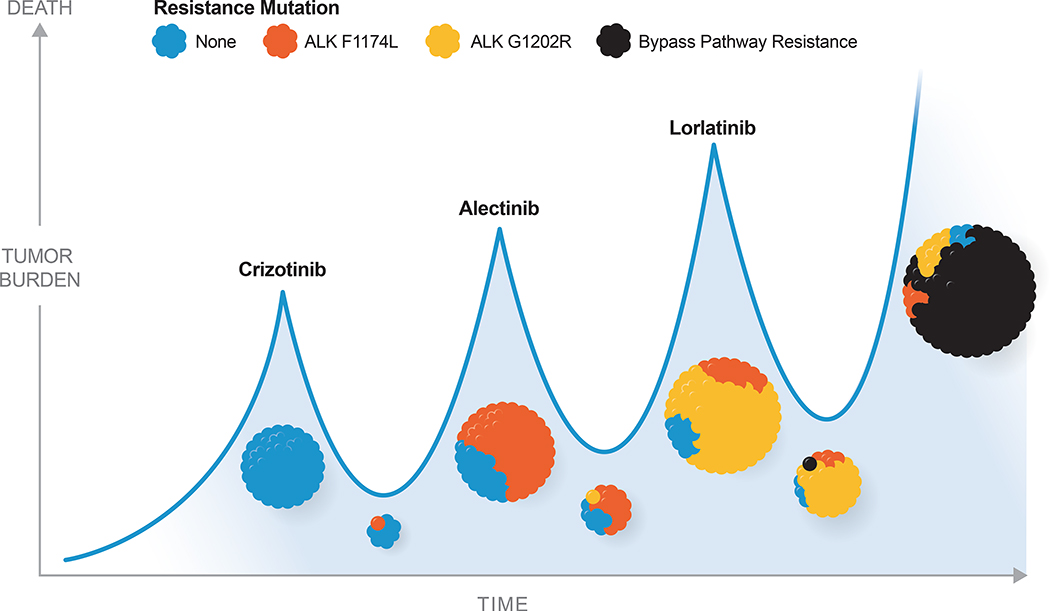
In addition to resistance liabilities dictated by anatomic drug penetrance, drug development has increasingly taken into account predicted mechanisms of on-target acquired resistance. Successive generations of ALK inhibitors, for example, have been designed specifically to maintain binding potency despite the acquisition of both individual and even compound mutations in the ALK kinase domain (Figure 2) (Crinò et al., 2016; Dagogo-Jack and Shaw, 2016; Gainor et al., 2016; Kim et al., 2016; McCusker et al., 2019; Ou et al., 2016; Shaw et al., 2016; Shaw et al., 2014; Zhang et al., 2016). On-target resistance has similarly proven a challenge in secondary resistance to multiple other genome-directed therapies, including those targeting EGFR (Balak et al., 2006; Kobayashi et al., 2005; Kosaka et al., 2006), ROS1 (Sehgal et al., 2018), NTRK (Schram et al., 2017a), and ABL, among others (Milojkovic and Apperley, 2009). Increasing access to highly sensitive tumor- and plasma-based sequencing platforms has created a feed-forward loop whereby the specific resistance liabilities of each successive generation of targeted therapy can be identified in real-time and drive design decisions of the next-generation inhibitor.
Figure 2. Schematic of ALK fusion-positive non-small cell lung cancer over time with exposure to sequential therapy.
Mutations in the ALK kinase domain that impair drug binding lead to acquired resistance in ALK fusion-positive NSCLC. Successive generations of ALK inhibitors have been designed to combat this on-target resistance and can be rationally sequenced in clinical practice to restore tumor control until a yet undruggable resistance mechanism emerges.
New Frontiers for Drug Development
Whereas small molecule inhibitors have generally inhibited both wild-type and mutant proteins, newer approaches have focused on specifically suppressing the activity of aberrant molecular targets using mutant and isoform-selective inhibitors. In parallel, breakthroughs in protein engineering have revealed new classes of agents capable of expanding the therapeutic index against validated drug targets, including antibody-drug conjugate (ADC) therapies that release cytotoxic payloads at the site of disease. Finally, entirely novel approaches initially pioneered in other areas of medicine, including protein-refolders that restore the function of pathogenic oncoproteins without inhibiting wildtype proteins, are now being explored in oncology.
Isoform and Mutant-Selective Inhibitors
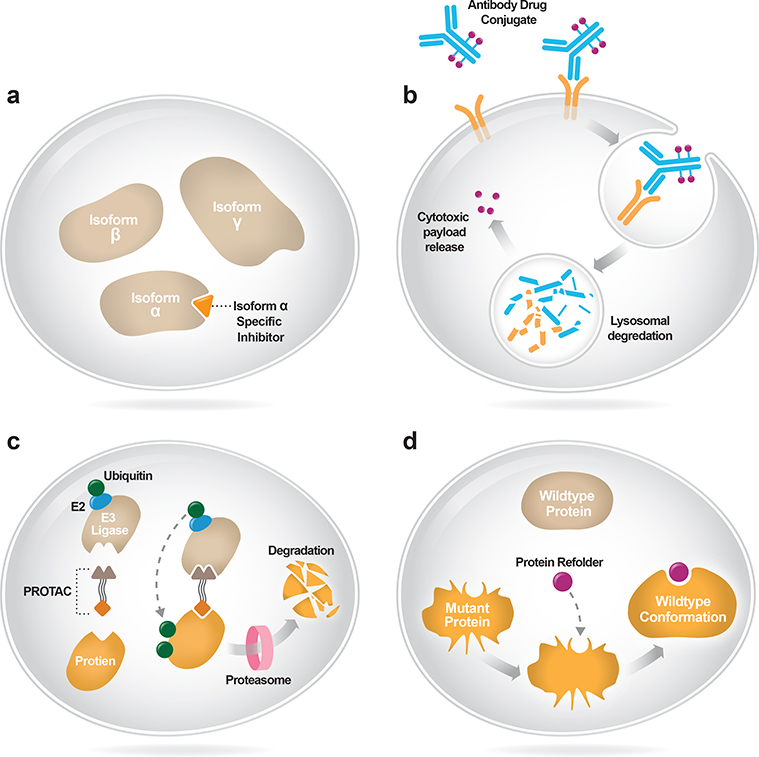
Recognizing that more selective therapies tend to have improved efficacy and tolerability, several strategies have been employed to more specifically and directly inhibit oncogenic drivers, including the development of isoform and mutant-selective therapies (Figure 3A). For example, the PI3K pathway is among the most commonly mutated in cancer (Thorpe et al., 2015; Zehir et al., 2017), but early efforts to target it with pan-PI3K inhibitors showed limited efficacy (Rodon et al., 2013). Isoform-selective PI3K inhibitors, by contrast, have shown improved outcomes relative to pan-PI3K and dual PI3K/mTOR inhibitors (Baselga et al., 2017; Yang et al., 2019). Moreover, isoform-specific inhibitors can be used to minimize toxicity attributable to an “off-target” isoform. Pan-FGFR inhibitors used to target FGFR2/3-altered cancers cause high rates of hyperphosphatemia, mediated predominantly by FGFR1 inhibition (Gattineni et al., 2014; Loriot et al., 2019; Nogova et al., 2017; Schram et al., 2017b; Wöhrle et al., 2011; Wu et al., 2013). Isoform-specific FGFR2 and/or FGFR3 inhibitors that spare FGFR1 are being developed to optimize tolerability and efficacy in FGFR2/3-altered tumors.
Figure 3A-D. New frontiers for drug development.
(A) Isoform-selective inhibitors bind to an individual protein isoform within the cell. (B) Antibody drug conjugates bind to cell surface antigens and are internalized into the cell where they release a cytotoxic payload to induce cell death. (C) Proteolysis Targeting Chimeras (PROTACs) bind both mutant proteins and E3 ubiquitin ligase, facilitating proteasomal degradation of the target. (D) Protein refolders enable mutant proteins to regain wildtype conformation and activity.
In recent years, drug selectivity has advanced beyond isoform selectivity and toward individual mutant alleles. Such selectivity allows for inhibition of the mutant oncogenic protein while sparing wild-type protein. In some cases, mutant allele selectivity may be the only way to pharmacologically inhibit an otherwise undruggable target. KRAS is among the most commonly mutated oncogenes in cancer (Mai and Lito, 2018; Ostrem et al., 2013; Zehir et al., 2017), but despite its recognition as a key oncogenic driver, it has historically be considered undruggable in part due to a lack of targetable binding pockets (Cox et al., 2014; McCormick, 2015; Stephen et al., 2014). Recently, however, improvements in small molecule design have facilitated the development of highly selective warheads that react with the mutant cysteine of KRAS G12C, forming an irreversible bond and locking the protein in its inactive GDP-bound state (Janes et al., 2018; Lito et al., 2016; Ostrem et al., 2013; Patricelli et al., 2016). In the absence of this mutant cysteine, these covalent inhibitors do not react with wildtype KRAS and thus spare host tissues. In keeping with the hypothesized wide therapeutic index of the drug based on mutant allele selectivity, early results from phase I trials of KRAS G12C inhibitors show minimal toxicity and early signs of efficacy in KRAS G12C-mutant NSCLC (Fakih et al., 2019; Jänne PA, 2019). Moreover, as mutant allele selective approaches such as KRAS G12C inhibitors have entered the clinic, the opportunity for combinatorial therapy has emerged and may finally permit the field to deliver on the promise of co-targeting cancer dependencies.
Antibody Drug Conjugates
Another approach to increasing the therapeutic index has been using antibody drug conjugates (ADCs). By directly linking a cytotoxic payload to a targeted antibody, ADCs are designed to expand the therapeutic window of traditional cytotoxic agents (Figure 3B) (Coats et al., 2019). Unfortunately, in many cases ADC toxicity has been greater than predicted for a variety of reasons, including host tissue expression of the target, non-specific cleavage of the toxin, and other less well understood mechanisms (Coats et al., 2019).
Through iterative improvement, the initial promise of this class of agents is finally beginning to deliver clinically. For example, the ADC trastuzumab deruxtecan (DS8201) is comprised of the anti-HER2 antibody trastuzumab conjugated to the cytotoxic topoisomerase I inhibitor, deruxtecan. This agent has shown unprecedented activity in HER2-driven cancers including HER2-amplified breast and gastric cancer (Modi et al., 2019; Shitara et al., 2019), in addition to promising activity in HER2-low breast cancer (Doi et al., 2017), a population for which HER2-targeted therapy has been largely ineffective (Modi et al., 2019; Shitara et al., 2019; Tamura et al., 2019). Identifying tumor-specific targets optimal for ADC development and optimizing the safety of these engineered drugs will be key to their further development and utilization.
Allosteric Inhibitors
Traditionally, the majority of small molecule inhibitors have targeted the ATP binding site. More recently, structure-based drug design, computational chemistry with dynamic simulation, and advances in high-throughput drug screening approaches have collectively enabled the development of non-ATP competitive inhibitors that engage novel allosteric sites. These allosteric inhibitors may overcome on-target resistance mediated by mutations in the active site of validated targets as well as facilitating inhibition of previously undruggable proteins. For example, the use of imatinib to target BCR-ABL fusion-positive CML was one of the first successes of precision medicine, often credited as launching the field (Longo, 2017). A series of successive generations of ATP-competitive ABL inhibitors followed, permitting salvage of an increasing variety of active site resistance mutations (O’Hare et al., 2007; O’Hare et al., 2009). However, these resistance mutations can become polyclonal and even develop in cis, limiting the effectiveness of kinase domain inhibitors (Deininger et al., 2016; Shah et al., 2002). For this reason, developing an orthogonal approach for inhibiting ABL through an allosteric mechanism not impacted by the acquisition of active site mutations is appealing. One such inhibitor, asciminib (ABL001), has already entered the clinic and demonstrated proof-of-concept in CML patients heavily pretreated with ATP-competitive inhibitors and harboring recalcitrant resistance mechanisms (Hughes et al., 2019; Wylie et al., 2017). Another approach being actively explored is the combination of selective ATP-competitive and allosteric inhibitors with non-overlapping resistance liabilities, which may collectively delay or even entirely forestall the development of acquired resistance.
Allosteric inhibitors may also enable therapy directed at previously undruggable targets. For example, the phosphatase SHP2, in conjunction with SOS1, plays an important role in enabling nucleotide exchange, allowing RAS to cycle between its inactive GDP-bound and its activated GTP-bound states (Chan and Feng, 2007; Grossmann et al., 2010; Lu et al., 2019; Mai and Lito, 2018; Mainardi et al., 2018; Matozaki et al., 2009; Nichols et al., 2018; Ostman et al., 2006; Ruess et al., 2018). Phosphatases had not previously been considered attractive drug targets, but allosteric inhibitors are in development that alter SHP2’s conformation and abrogate its activity, leading to inhibition of MAPK-driven tumors in preclinical models and prompting several ongoing clinical trials (Frankson et al., 2017; Mai and Lito, 2018).
Proteolysis Targeting Chimeras (PROTACS)
Another emerging approach to targeting key cancer dependencies relies on protein degraders, referred to by a variety of terms including Proteolysis Targeting Chimeras (PROTACS) and “molecular glues”, among other names. This heterogeneous group of therapies generally involve the use of a bifunctional molecule that brings the intended target into the proximity of a ubiquitin ligase, ultimately leading to the target’s degradation (Figure 3C). The application of this technology to cancer therapeutics is still in its infancy, but is potentially multifaceted. Like allosteric inhibitors of key oncogenes, this technique may degrade key cancer dependencies even in the setting of resistance mutations that otherwise render existing therapeutics inactive. Perhaps most exciting is the ability this technology possesses to engage proteins without catalytic sites, those that lack traditional deep binding pockets required of kinase inhibitors, or otherwise difficult to drug transcription factors. Many key drivers of cancer, including transcription factors, cannot be targeted by currently available therapeutics, either because they are not expressed on the cell surface and therefore are inaccessible to antibodies or because they lack a binding pocket to which a small molecule inhibitor can attach. PROTACs may overcome these challenges and exploit the endogenous protein degradation machinery of the cell by simultaneously binding a target and the E3 ubiquitin ligase, promoting protein degradation (Chamberlain and Hamann, 2019; Schapira et al., 2019). ARV-110 is the first such drug to enter phase I clinical trials and links the E3 ubiquitin ligase with the androgen receptor in patients with prostate cancer (NCT03888612). This novel approach to decrease cellular protein levels may make it possible to effectively target numerous previously undruggable targets.
Protein Refolders
Small molecules are being developed to restore the natural function of mutant proteins by molding protein conformation to re-enable lost activity (Figure 3D). This strategy has already proven successful in the treatment of cystic fibrosis, a non-oncologic hereditary disease characterized by excessive mucus production due to mutations in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR) protein. By re-enabling CFTR to reach the cell surface and function similarly to wildtype protein, protein refolders diminish the clinical sequelae of cystic fibrosis (Middleton et al., 2019). The application of protein refolders to cancer is currently being explored and represents a novel means of targeting mutant tumor suppressors. Loss-of-function mutations in the tumor suppressor TP53 are the most common mutations in cancer (Zehir et al., 2017). However, there are currently no therapies approved that specifically target TP53-altered cancers. Efforts are underway to develop small molecules that restore the activity of mutant TP53 through protein refolding (Blandino and Di Agostino, 2018; Bykov et al., 2002). In addition to expanding the number of possible drug targets, this approach provides the additional benefit of being mutant specific, thereby decreasing toxicity.
Improved Patient Selection
Along with optimizing the drugs brought to clinic, genome-driven oncology is evolving to improve patient selection for clinical trials. This refined patient matching is enabled by our growing understanding of the interaction between specific molecular alterations and the histologic and broader genomic context of the tumor as well as our improved ability to recognize oncogenic drivers among a much larger number of variants of unknown significance in individual tumors.
Histology Matters (Sometimes)
There is increasing recognition of the value of histology agnostic molecularly-matched therapies. This is exemplified by TRK inhibitors, which exhibit potent antitumor activity in TRK-fusion positive cancers regardless of tumor histology (Cocco et al., 2018; Doebele et al., 2020; Drilon et al., 2018b; Liu et al., 2018). The success of these drugs led to TRK selective therapies being the first molecularly targeted treatments approved by regulatory agencies for use across all solid tumors.
While TRK inhibition drives tumor-agnostic efficacy, the activity of most targeted therapies is conditioned by the tumor histology. For example, combination BRAF and MEK inhibition has become standard first-line therapy for BRAF V600E-mutant melanoma and non-small cell lung cancers, but is only minimally active in colorectal cancer (Corcoran et al., 2015; Dummer et al., 2018a; Dummer et al., 2018b; Planchard et al., 2016). Similarly, HER2-mutant tumors treated with the pan-HER kinase inhibitor neratinib demonstrate marked differences in sensitivity based on tumor lineage with higher response rates among patients with breast, cervical, and biliary cancers (Hyman et al., 2018). The importance of histology also holds true for certain targetable germline alterations. In a retrospective analysis of BRCA-mutant patients, tumor lineage was the most important predictor of benefit from PARP inhibition (Jonsson et al., 2019). Basket studies exploring the efficacy of targeted therapy across histologies are useful in identifying the characteristics of patients most likely to benefit from a given therapy and serve as an initial step in developing genomically-targeted treatments in the appropriate histologic context.
Genomic Context Matters
While most targeted therapies are designed to inhibit a single driver mutation, co-alterations may impact the efficacy of these therapies (Huang et al., 2020). For example, in patients with HER2-mutant metastatic breast cancer, concurrent mutations in HER2 or HER3 have been associated with a decreased likelihood of response to neratinib (Smyth et al., 2019). Similarly, in patients with AKT1 E17K mutations treated with a pan-AKT inhibitor, those with concurrent alterations in the PI3K pathway had shorter progression free survival (PFS) than those without these co-mutations (Hyman et al., 2017a). In patients with EGFR-mutant NSCLC, MET and ERBB2 amplification have been reported to cause primary resistance to third-generation EGFR inhibitors, including osimertinib (Ortiz-Cuaran et al., 2016). While co-mutations in alternative drivers are associated with decreased likelihood of response to targeted therapies, they do not eliminate the possibility of responses in some patients. Furthermore, it is not always possible to predict whether a co-mutation is a second driver or merely a passenger mutation. Therefore, further research is needed to determine optimal matching of patients to drugs informed by the co-mutational context.
Features of the targeted mutation itself, including whether it represents a truncal or branch alteration, may also affect the likelihood of response to therapy. For example, the relatively large proportion of sub-clonal PIK3CA mutations may add to the complexity of PIK3CA inhibition (Amirouchene-Angelozzi, et al., 2017; McGranahan, et al., 2015). While high variant allele frequency may provide supportive evidence for the driver status of an alteration (Spurr, et al., 2018), patients with low allele frequency alterations have also responded to targeted therapies (Shin, et al., 2017). Optimal thresholds for defining targetable amplifications are also still under investigation and may vary based on the gene and therapeutic agent (Lai, et al., 2019).
Zygosity has also emerged as another potential mediator of responsiveness to targeted therapies. In patients with AKT1 E17K mutations treated with AZD5363, mutant allele imbalance most often caused by loss of WT AKT1 resulted in a prolonged PFS compared to patients with heterozygous AKT1 E17K tumors (Hyman et al., 2017a). Similarly, in patients with BRAF V600-mutant melanoma, loss of the wild-type allele resulted in improved PFS compared to patients whose tumors were heterozygous at this allele. Interestingly, patients with genomic gains of mutant BRAF V600 did not demonstrate improved outcomes compared to those with heterozygous BRAF V600, implying that loss of the wild-type allele may be more important than the dose of mutant oncogene in sensitizing to targeted therapy (Bielski et al., 2018). By contrast, the zygosity of BRCA1/2 mutations did not mediate responsiveness to PARP inhibition in a retrospective analysis of BRCA-mutant patients, suggesting that the implications of zygosity for responsiveness may vary depending on the biomarker being targeted and the affected tumor lineage (Jonsson et al., 2019).
Triaging Variants of Unknown Significance
A critical component of precision oncology is identifying which patients harbor druggable targets. A major challenge has been characterizing which variants of unknown significance (VUSs) found by broad next-generation sequencing actually drive tumor growth. Computational analyses of population-scale genomic data have identified novel “hotspot” mutations that are altered more frequently in cancer than would be predicted in the absence of selection. This positive selection suggests that hotspot mutations may be oncogenic and functionally important (Chang et al., 2018; Koyama et al., 2019). As proof-of-principle, several patients with VUSs in targets for which investigational therapies existed were enrolled on relevant therapies and benefited clinically based solely on the knowledge that their mutation was a hotspot (Chang et al., 2018). While not every hotspot is functional (Buisson et al., 2019), such computational weight of evidence is an important tool for identifying still occult driver mutations. Numerous additional methods are aiding in further characterizing variants of unknown significance, including saturation mutagenesis in genes of interest (Findlay et al., 2014) and multiplexed functional assays (Gasperini et al., 2016).
Optimizing the use of current and future drugs
To maximize the benefit of genome-driven oncology, it is essential to optimize the use of existing therapies. Giving novel therapies at the most appropriate time within a patients’ treatment course may enhance outcomes. Rational sequencing of treatments and the development of synergistic and tolerable combinations are two mechanisms by which physicians can improve patient outcomes using existing tools.
Timing Therapies to Minimize Resistance
Novel therapies are typically tested in patients with disease that has received maximum benefit from existing standard treatments. However, the experience with EGFR and ALK inhibitors suggests that outcomes may be improved with earlier application of our best drugs, prior to the development of resistance. Over 50% of EGFR-mutant NSCLC patients treated with first- and second-generation EGFR tyrosine kinase inhibitors (TKIs) acquire EGFR T790M mutations (Oxnard et al., 2011; Yu et al., 2013). Osimertinib was developed to overcome the T790M mutation and was initially studied in patients who had progressed on prior TKIs (Mok et al., 2017). More recent evidence demonstrates that when patients receive osimertinib as first-line therapy, effectively preventing T790M-mediated resistance, overall survival is significantly increased compared to patients treated with first-generation EGFR-TKIs (Ramalingam et al., 2020). Drug development paradigms should therefore encourage the testing of later generation inhibitors upfront.
Adjuvant and Neoadjuvant Therapy
The majority of genome-driven therapy is administered to patients with recurrent or metastatic cancer where the goal is to prolong life without the expectation of cure. Conversely, the greatest opportunity for targeted therapy may instead be in patients with earlier stage disease where effective therapies have the potential to increase cure rates. In patients with HER2-positive breast cancer, the addition of the HER2 monoclonal antibody trastuzumab to chemotherapy resulted in a substantial increase in 10-year survival (Perez et al., 2014). Adjuvant targeted therapy is also standard of care for patients with stage 3 BRAF V600E-mutant melanoma and KIT-expressing gastrointestinal stromal tumors greater than 3 centimeters in size based on phase 3 trials that demonstrated disease-free survival benefits (Dematteo et al., 2009; Long et al., 2017). For targeted therapies with a high response rate, neoadjuvant therapy can be utilized to convert unresectable tumors to surgically resectable disease, thereby providing an opportunity for cure. For example, although larotrectinib was developed for advanced TRK fusion-positive cancers unresponsive to standard treatments, neoadjuvant larotrectinib has been successfully used in pediatric sarcoma patients to shrink tumors and allow for complete resections (Drilon et al., 2018b; DuBois et al., 2018). While further research is needed to define when such a neoadjuvant approach is appropriate, it is clear that earlier use of effective drugs may dramatically improve prognosis in a subset of patients.
Rational Combinations
Combination therapy can be utilized to increase efficacy, decrease toxicity, and/or prevent the emergence of resistance. In BRAF V600-mutant melanoma, the combination of BRAF and MEK inhibition results in prolonged survival and decreased cutaneous toxicity (squamous-cell carcinomas and keratoacanthomas) compared to BRAF inhibition alone (Kopetz et al., 2019; Long et al., 2014; Robert et al., 2015). This improved toxicity profile is made possible by the unique properties of BRAF inhibitors which target the BRAF V600 monomer in cancer cells, but paradoxically transactivate the wildtype kinase in host tissues, creating a therapeutic index (Su et al., 2012). In colon cancers harboring BRAF V600 mutations, however, RAF inhibition causes rapid feedback activation through EGFR and triple therapy with BRAF, MEK, and EGFR inhibition is required to achieve a meaningful clinical benefit (Kopetz et al., 2019). Thus, the most effective combinations may depend not only on the genomic biomarker of interest but also on the tumor type being treated.
In addition to preventing primary resistance, rational combinations can effectively treat secondary resistance. Off-target resistance has been increasingly reported as post-progression biopsies become commonplace. When the acquired alteration is itself targetable, sequencing data provides the opportunity for rational therapeutic combinations that simultaneously target the primary and acquired drivers (Cocco et al., 2019; Sequist et al., 2019). In the TATTON trial, patients with EGFR-mutant NSCLC with acquired MET amplifications were treated with osimertinib in combination with the MET inhibitor savolitinib. Savolitinib was able to restore sensitivity to osimertinib and was associated with radiographic responses (Sequist et al., 2019). Importantly, the clinical and therapeutic relevance of MET copy number gains and amplification likely depends on the technique used to measure this and standard definitions must be adopted and relevant thresholds defined in order to optimize this biomarker for clinical use (Lai, et al., 2019).
Next-generation Clinical Studies
Another frontier in advancing precision oncology involves novel clinical trial designs that assign therapies based on an individual patients’ tumor genomics or adapt therapy based on prognostic biomarkers. Historically clinical trials enrolled patients with a specific cancer type and stage of disease and offered patients either predetermined therapy or randomization to one of several study arms. As understanding of biomarkers for responsiveness and resistance to therapy has grown, so too have the list of strategies used to investigate treatment. We must continue to refine clinical trial designs to more precisely match individual patients to appropriate treatments.
Genomic Matching: Beyond the Basket Trial
Clinical trials utilizing genomic biomarkers were initially carried out in single tumor types. For example, an investigation of PARP inhibitors in heavily pre-treated prostate cancer patients demonstrated efficacy in individuals with alterations in the DNA damage repair genes BRCA1/2 and ATM (Mateo, et al., 2015). Recently, basket trials that enroll patients with a biomarker regardless of tumor type have become more widespread (Tao et al., 2018). This design allows for the investigation of rare genomic alterations and inclusion of rare tumor types that could not be practically studied in a more traditional model. Umbrella and platform studies are also emerging whereby patients with a particular tumor type are enrolled to treatment arms based on their underlying genomic profile (West, 2017). Platform studies build in the potential for iteration and addition of novel therapies as these become available (Adaptive Platform Trials Coalition, 2019).
Master protocols such as NCI-MATCH allow patients across disease types to be treated with a number of genomically-matched therapies. NCI-MATCH demonstrated the feasibility of this approach on a large scale and highlighted the high number of patients interested in genomically-directed trials, however, accrual was initially lower than predicted. This was due to several factors including the frequency of actionability, the availability of approved drugs for identified alterations making patients ineligible, the turn-around-time for genomic sequencing, and need for sufficient tissue. Rapid molecular profiling, tumor sequencing earlier in a patient’s disease course, and education regarding acceptable tissue samples will be critical for future large-scale precision oncology initiatives (Flaherty et al., 2020).
Innovative trial designs have allowed for more efficient testing of biomarker-driven hypotheses. Building on these principles, future clinical trial designs will continue to assign patients to treatments based on ever more specific and individualized factors, such as mechanisms of acquired resistance to prior therapy. In the ORCHARD trial (Osimertinib Resistance Cohorts Addressing 1L Relapse Drivers), patients with EGFR-mutant NSCLC who have progressed on first-generation TKIs undergo pretreatment biopsies and are stratified to osimertinib in combination with another agent based on the mechanism of acquired resistance (or lack thereof) (NCT 03944772). This trial design allows for streamlined testing of multiple rational combination therapies.
Adaptation of therapy based on dynamic tumor monitoring
Collection of cell-free DNA (cfDNA) shed from tumors is emerging as a means to dynamically monitor response to therapy or tumor recurrence after definitive treatment and can be used for risk stratification and potentially adapting therapy in creative clinical trial designs. In a trial of AKT-mutant tumors treated with an AKT inhibitor, a cfDNA decline of 50% at day 21 was associated with improved PFS (Hyman et al., 2017a). Similarly, a retrospective analysis of patients with breast cancer treated with combined therapy consisting of the CDK4/6 inhibitor palbociclib with the selective estrogen receptor degrader fulvestrant demonstrated that patients with a cfDNA decline on treatment had prolonged (O’Leary et al., 2018). In women with early stage breast cancer, the detection of cfDNA after initial treatment is also associated with increased risk of recurrence (Garcia-Murillas et al., 2019). Thus, cfDNA may have utility as a predictive and prognostic marker to study treatment de-escalation in low-risk patients and/or intensification in high-risk patients on treatment or after definitive surgery. In an ongoing study of patients with solid tumors demonstrating microsatellite instability, patients with detectable cfDNA after definitive treatment are randomized to either placebo or treatment intensification with adjuvant pembrolizumab to determine whether treating microscopic disease with immunotherapy prolongs survival (NCT 03832569). The success of such trials utilizing cfDNA may change the way we monitor cancer and redefine disease persistence and recurrence.
Capitalizing on Real World Data
As precision oncology turns its attention to the many rare genomic alterations that drive cancer, obtaining reliable information on individual genomic phenotypes will increasingly depend on collecting real world data (RWD) from patients treated on and off trials (AACR Project GENIE Consortium, 2017; Armenia et al., 2018; Chang et al., 2018; Dickson et al., 2020; Hyman et al., 2017b). Several initiatives have been launched with the aim of capturing RWD. The American Association for Cancer Research (AACR) has created Project GENIE (Genomics Evidence Neoplasia Information Exchange), which serves as a clinical and genomic data repository, collating information from across multiple centers nationally (AACR Project GENIE Consortium, 2017). GENIE has already been used to characterize the natural history of AKT1 E17K-mutant, estrogen receptor positive breast cancer. American Society of Clinical Oncology (ASCO) has launched CancerLinQ (Cancer Learning Intelligence Network for Quality), which uses the electronic health records of participating institutions to explore issues such as optimal drug sequencing and the impact of comorbid conditions on treatment toxicity (Rubinstein and Warner, 2018). Similarly, various industry partners are creating platforms for assembling data from the electronic medical records of patients treated in both academic and community forums, allowing for large-scale retrospective investigations of outcomes data (Khozin et al., 2019; Moser et al., 2020; Parikh et al., 2019).
Regulatory agencies have recognized the importance of collecting information on patients treated outside of clinical trials. As clinical trials have increasingly focused on rare genomic alterations and diseases, regulatory agencies have acknowledged that large randomized controlled trials may not be feasible in all circumstances and single-arm studies that demonstrate unequivocal efficacy may be sufficient for drug approval. In this circumstance, evaluating RWD post-approval is critical to confirm improvement in patient outcomes. The FDA’s Real World Evidence program was created following a congressional mandate as part of the 21st Century Cures Act passed in 2016. The program has focused on using RWD not only for post-approval investigations, but also to accelerate the expansion of drug approvals for new indications (U.S. Food and Drug Administration, 2018). The approval of palbociclib, a CDK4/6 inhibitor, was recently expanded from women with HER2-negative metastatic breast cancer to also include male breast cancer based in part on RWD (Wedam et al., 2019). As RWD increasingly supplements clinical trial data, standardization of data collection and usage will be critical.
Conclusion
Tumor molecular profiling has enabled the development of highly successful targeted therapies that have benefitted countless patients. Yet molecularly-directed studies have also underscored the complexity of predicting which patients will respond to therapy. Factors ranging from co-alterations in the tumor itself to lineage specificity alter patients’ likelihood of responding to today’s treatments. Moreover, many genomic drivers remain “undruggable” or are ineffectively targeted due to poor tolerability of current therapies. To realize the promise of genome-directed therapy, we must learn from prior successes and failures to optimize drug design, develop innovative new therapeutic approaches, and refine how patients are matched to treatment.
Acknowledgments
The authors gratefully acknowledge Scott Johnson and Christopher Kaeser for their help with graphics support and funding from the National Cancer Institute (NCI) under the MSKCC Support Grant/Core Grant (P30 CA008748).
Declaration of Interests:
YRMG acknowledges receipt of support for travel, accommodation and expenses from AstraZeneca and has received training through a K30 grant (CTSA UL1TR00457). B.S.T. reports receiving honoraria and research funding from Genentech and Illumina and advisory board activities for Boehringer Ingelheim and Loxo Oncology, a wholly owned subsidiary of Eli Lilly. DMH reports paid consulting for Chugai, Boehringer Ingelheim, AstraZeneca, Pfizer, Bayer, Genentech/Roche, Fount Therapeutics, and research grants from AstraZeneca, Puma Biotechnology, Loxo Oncology, Bayer. He is currently employed by Loxo, a wholly owned subsidiary of Eli Lilly. AMS has no financial interests to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.AACR Project GENIE Consortium (2017). AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 7, 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann CJ, Stock G, Tay R, Dawod M, Gomes F, and Califano R (2019). Targeted therapy for RET-rearranged non-small cell lung cancer: clinical development and future directions. OncoTargets Ther. 12, 7857–7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adaptive Platform Trials Coalition (2019). Adaptive platform trials: definition, design, conduct and reporting considerations. Nat. Rev. Drug Discov. 18, 797–807. [DOI] [PubMed] [Google Scholar]
- 4.Amirouchene-Angelozzi N, Swanton C, Bardelli A (2017). Tumor evolution as a therapeutic target. Cancer Discov 7, 1–13. [DOI] [PubMed] [Google Scholar]
- 5.Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, Chatila WK, Chakravarty D, Han GC, Coleman I, et al. (2018). The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azad NS, Aragon-Ching JB, Dahut WL, Gutierrez M, Figg WD, Jain L, Steinberg SM, Turner ML, Kohn EC, and Kong HH (2009). Hand-foot skin reaction increases with cumulative sorafenib dose and with combination anti-vascular endothelial growth factor therapy. Clin. Cancer Res. 15, 1411–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, Chiang A, Yang G, Ouerfelli O, Kris MG, et al. (2006). Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin. Cancer Res. 12, 6494–6501. [DOI] [PubMed] [Google Scholar]
- 8.Baselga J, Im S-A, Iwata H, Cortés J, De Laurentiis M, Jiang Z, Arteaga CL, Jonat W, Clemons M, Ito Y, et al. (2017). Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beau-Faller M, Prim N, Ruppert AM, Nanni-Metéllus I, Lacave R, Lacroix L, Escande F, Lizard S, Pretet JL, Rouquette I, et al. (2014). Rare EGFR exon 18 and exon 20 mutations in non-small-cell lung cancer on 10 117 patients: a multicentre observational study by the French ERMETIC-IFCT network. Ann. Oncol. 25, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielski CM, Donoghue MTA, Gadiya M, Hanrahan AJ, Won HH, Chang MT, Jonsson P, Penson AV, Gorelick A, Harris C, et al. (2018). Widespread Selection for Oncogenic Mutant Allele Imbalance in Cancer. Cancer Cell 34, 852–862.e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blandino G, and Di Agostino S (2018). New therapeutic strategies to treat human cancers expressing mutant p53 proteins. J. Exp. Clin. Cancer. Res. 37, 30–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandhuber BB, H. J., Tuch BB, et al. , ENA-0490 The development of LOXO-292, a potent, KDR/VEGFR2-sparing RET kinase inhibitor for treating patients with RET-dependent cancers. In AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics, (Munich, Germany ). [Google Scholar]
- 13.Buisson R, Langenbucher A, Bowen D, Kwan EE, Benes CH, Zou L, and Lawrence MS (2019). Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science 364, eaaw2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bykov VJN, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, and Selivanova G (2002). Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 8, 282–288. [DOI] [PubMed] [Google Scholar]
- 15.Camidge DR, Kim HR, Ahn M-J, Yang JC-H, Han J-Y, Lee J-S, Hochmair MJ, Li JY-C, Chang G-C, Lee KH, et al. (2018). Brigatinib versus Crizotinib in ALK-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 379, 2027–2039. [DOI] [PubMed] [Google Scholar]
- 16.Chamberlain PP, and Hamann LG (2019). Development of targeted protein degradation therapeutics. Nat. Chem. Biol. 15, 937–944. [DOI] [PubMed] [Google Scholar]
- 17.Chan RJ, and Feng G-S (2007). PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood 109, 862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang MT, Bhattarai TS, Schram AM, Bielski CM, Donoghue MTA, Jonsson P, Chakravarty D, Phillips S, Kandoth C, Penson A, et al. (2018). Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov. 8, 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chouitar J, Vincent S, Brake R, and Li S (2018). [Abstract] P2.13–32 TAK-788 is a Novel and Potent Tyrosine Kinase Inhibitor with Selective Activity Against EGFR/HER2. J. Thor. Oncol. 13, S811. [Google Scholar]
- 20.Coats S, Williams M, Kebble B, Dixit R, Tseng L, Yao N-S, Tice DA, and Soria J-C (2019). Antibody-Drug Conjugates: Future Directions in Clinical and Translational Strategies to Improve the Therapeutic Index. Clin. Cancer Res. 25, 5441–5448. [DOI] [PubMed] [Google Scholar]
- 21.Cocco E, Scaltriti M, and Drilon A (2018). NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 15, 731–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cocco E, Schram AM, Kulick A, Misale S, Won HH, Yaeger R, Razavi P, Ptashkin R, Hechtman JF, Toska E, et al. (2019). Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat. Med. 25, 1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corcoran RB, Atreya CE, Falchook GS, Kwak EL, Ryan DP, Bendell JC, Hamid O, Messersmith WA, Daud A, Kurzrock R, et al. (2015). Combined BRAF and MEK Inhibition With Dabrafenib and Trametinib in BRAF V600-Mutant Colorectal Cancer. J. Clin. Oncol. 33, 4023–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox AD, Fesik SW, Kimmelman AC, Luo J, and Der CJ (2014). Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13, 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crinò L, Ahn M-J, De Marinis F, Groen HJM, Wakelee H, Hida T, Mok T, Spigel D, Felip E, Nishio M, et al. (2016). Multicenter Phase II Study of Whole-Body and Intracranial Activity With Ceritinib in Patients With ALK-Rearranged Non-Small-Cell Lung Cancer Previously Treated With Chemotherapy and Crizotinib: Results From ASCEND-2. J. Clin. Oncol. 34, 2866–2873. [DOI] [PubMed] [Google Scholar]
- 26.Dagogo-Jack I, and Shaw AT (2016). Crizotinib resistance: implications for therapeutic strategies. Ann. Oncol. 27 Suppl 3, iii42–iii50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deininger MW, Hodgson JG, Shah NP, Cortes JE, Kim D-W, Nicolini FE, Talpaz M, Baccarani M, Müller MC, Li J, et al. (2016). Compound mutations in BCR-ABL1 are not major drivers of primary or secondary resistance to ponatinib in CP-CML patients. Blood 127, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dematteo RP, Ballman KV, Antonescu CR, Maki RG, Pisters PWT, Demetri GD, Blackstein ME, Blanke CD, von Mehren M, Brennan MF, et al. (2009). Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet 373, 1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickson D, Johnson J, Bergan R, Owens R, Subbiah V, and Kurzrock R (2020). The Master Observational Trial: A New Class of Master Protocol to Advance Precision Medicine. Cell 180, 9–14. [DOI] [PubMed] [Google Scholar]
- 30.Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, Blakely CM, Seto T, Cho BC, Tosi D, et al. (2020). Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol. 21, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doi T, Shitara K, Naito Y, Shimomura A, Fujiwara Y, Yonemori K, Shimizu C, Shimoi T, Kuboki Y, Matsubara N, et al. (2017). Safety, pharmacokinetics, and antitumour activity of trastuzumab deruxtecan (DS-8201), a HER2-targeting antibody-drug conjugate, in patients with advanced breast and gastric or gastro-oesophageal tumours: a phase 1 dose-escalation study. Lancet Oncol. 18, 1512–1522. [DOI] [PubMed] [Google Scholar]
- 32.Drilon A, Fu S, Patel MR, Fakih M, Wang D, Olszanski AJ, Morgensztern D, Liu SV, Cho BC, Bazhenova L, et al. (2019). A Phase I/Ib Trial of the VEGFR-Sparing Multikinase RET Inhibitor RXDX-105. Cancer Discov. 9, 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drilon A, Hu ZI, Lai GGY, and Tan DSW (2018a). Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nat. Rev. Clin. Oncol. 15, 151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, Nathenson M, Doebele RC, Farago AF, Pappo AS, et al. (2018b). Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med. 378, 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drilon A, Rekhtman N, Arcila M, Wang L, Ni A, Albano M, Van Voorthuysen M, Somwar R, Smith RS, Montecalvo J, et al. (2016). Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol. 17, 1653–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drilon AE, Subbiah V, Oxnard GR, Bauer TM, Velcheti V, Lakhani NJ, Besse B, Park K, Patel JD, Cabanillas ME, et al. (2018c). [Abstract] A phase 1 study of LOXO-292, a potent and highly selective RET inhibitor, in patients with RET-altered cancers. J. Clin. Oncol. 36, 102–102. [Google Scholar]
- 37.DuBois SG, Laetsch TW, Federman N, Turpin BK, Albert CM, Nagasubramanian R, Anderson ME, Davis JL, Qamoos HE, Reynolds ME, et al. (2018). The use of neoadjuvant larotrectinib in the management of children with locally advanced TRK fusion sarcomas. Cancer 124, 4241–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dummer R, Ascierto PA, Gogas HJ, Arance A, Mandala M, Liszkay G, Garbe C, Schadendorf D, Krajsova I, Gutzmer R, et al. (2018a). Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 19, 603–615. [DOI] [PubMed] [Google Scholar]
- 39.Dummer R, Ascierto PA, Gogas HJ, Arance A, Mandala M, Liszkay G, Garbe C, Schadendorf D, Krajsova I, Gutzmer R, et al. (2018b). Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 19, 1315–1327. [DOI] [PubMed] [Google Scholar]
- 40.Dvorakova S, Vaclavikova E, Sykorova V, Vcelak J, Novak Z, Duskova J, Ryska A, Laco J, Cap J, Kodetova D, et al. (2008). Somatic mutations in the RET proto-oncogene in sporadic medullary thyroid carcinomas. Mol. Cell. Endocrinol. 284, 21–27. [DOI] [PubMed] [Google Scholar]
- 41.Elisei R, Cosci B, Romei C, Bottici V, Renzini G, Molinaro E, Agate L, Vivaldi A, Faviana P, Basolo F, et al. (2008). Prognostic significance of somatic RET oncogene mutations in sporadic medullary thyroid cancer: a 10-year follow-up study. J. Clin. Endocrinol. Metab. 93, 682–687. [DOI] [PubMed] [Google Scholar]
- 42.Elisei R, Schlumberger MJ, Muller SP, Schoffski P, Brose MS, Shah MH, Licitra L, Jarzab B, Medvedev V, Kreissl MC, et al. (2013). Cabozantinib in progressive medullary thyroid cancer. J. Clin. Oncol. 31, 3639–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fakih M, O’Neil B, Price TJ, Falchook GS, Desai J, Kuo J, Govindan R, Rasmussen E, Morrow PKH, Ngang J, et al. (2019). [Abstract] Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J. Clin. Oncol. 37, 3003–3003. [Google Scholar]
- 44.Findlay GM, Boyle EA, Hause RJ, Klein JC, and Shendure J (2014). Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513, 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flaherty KT, Gray R, Chen A, Li S, Patton D, Hamilton SR, Williams PM, Mitchell EP, Iafrate AJ, Sklar J, et al. (2020). The Molecular Analysis for Therapy Choice (NCI-MATCH) Trial: Lessons for Genomic Trial Design J. Natl. Cancer. Inst, djz245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frankson R, Yu Z-H, Bai Y, Li Q, Zhang R-Y, and Zhang Z-Y (2017). Therapeutic Targeting of Oncogenic Tyrosine Phosphatases. Cancer Res. 77, 5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gadgeel S, Peters S, Mok T, Shaw AT, Kim DW, Ou SI, Pérol M, Wrona A, Novello S, Rosell R, et al. (2018). Alectinib versus crizotinib in treatment-naive anaplastic lymphoma kinase-positive (ALK+) non-small-cell lung cancer: CNS efficacy results from the ALEX study. Ann. Oncol. 29, 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, Dagogo-Jack I, Gadgeel S, Schultz K, Singh M, et al. (2016). Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer . Cancer Discov. 6, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gainor JF, Lee DH, Curigliano G, Doebele RC, Kim D-W, Baik CS, Tan DS-W, Lopes G, Gadgeel SM, Cassier PA, et al. (2019). [Abstract] Clinical activity and tolerability of BLU-667, a highly potent and selective RET inhibitor, in patients (pts) with advanced RET-fusion+ non-small cell lung cancer (NSCLC). J. Clin. Oncol. 37, 9008–9008. [Google Scholar]
- 50.Garcia-Murillas I, Chopra N, Comino-Méndez I, Beaney M, Tovey H, Cutts RJ, Swift C, Kriplani D, Afentakis M, Hrebien S, et al. (2019). Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol, e191838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gasperini M, Starita L, and Shendure J (2016). The power of multiplexed functional analysis of genetic variants. Nat. Protoc. 11, 1782–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gattineni J, Alphonse P, Zhang Q, Mathews N, Bates CM, and Baum M (2014). Regulation of renal phosphate transport by FGF23 is mediated by FGFR1 and FGFR4. Am. J. Physiol. Renal Physiol. 306, F351–F358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gautschi O, Milia J, Filleron T, Wolf J, Carbone DP, Owen D, Camidge R, Narayanan V, Doebele RC, Besse B, et al. (2017). Targeting RET in Patients With RET-Rearranged Lung Cancers: Results From the Global, Multicenter RET Registry. J. Clin. Oncol. 35, 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grossmann KS, Rosário M, Birchmeier C, and Birchmeier W (2010). The tyrosine phosphatase Shp2 in development and cancer. Adv. Cancer Res. 106, 53–89. [DOI] [PubMed] [Google Scholar]
- 55.Guo R, Schreyer M, Chang JC, Rothenberg SM, Henry D, Cotzia P, Kris MG, Rekhtman N, Young RJ, Hyman DM, and Drilon A (2019). Response to Selective RET Inhibition With LOXO-292 in a Patient With RET Fusion-Positive Lung Cancer With Leptomeningeal Metastases . JCO Precis. Oncol. 3, 10.1200/PO.1219.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han J-Y, Lee KH, Kim S-W, Min YJ, Cho E, Lee Y, Lee S-H, Kim HY, Lee GK, Nam BH, et al. (2017). A Phase II Study of Poziotinib in Patients with Epidermal Growth Factor Receptor (EGFR)-Mutant Lung Adenocarcinoma Who Have Acquired Resistance to EGFR-Tyrosine Kinase Inhibitors. Cancer Res. Treat. 49, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayman SR, Leung N, Grande JP, and Garovic VD (2012). VEGF inhibition, hypertension, and renal toxicity. Current oncology reports 14, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang A, Garraway LA, Ashworth A, and Weber B (2020). Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 19, 23–38. [DOI] [PubMed] [Google Scholar]
- 59.Hughes TP, Mauro MJ, Cortes JE, Minami H, Rea D, DeAngelo DJ, Breccia M, Goh Y-T, Talpaz M, Hochhaus A, et al. (2019). Asciminib in Chronic Myeloid Leukemia after ABL Kinase Inhibitor Failure. N. Engl. J. Med. 381, 2315–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, Juric D, Quinn DI, Moreno V, Doger B, et al. (2018). HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 554, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hyman DM, Smyth LM, Donoghue MTA, Westin SN, Bedard PL, Dean EJ, Bando H, El-Khoueiry AB, Pérez-Fidalgo JA, Mita A, et al. (2017a). AKT Inhibition in Solid Tumors With AKT1 Mutations. J. Clin. Oncol. 35, 2251–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hyman DM, Taylor BS, and Baselga J (2017b). Implementing Genome-Driven Oncology. Cell 168, 584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janes MR, Zhang J, Li L-S, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, et al. (2018). Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 172, 578–589.e517. [DOI] [PubMed] [Google Scholar]
- 64.Jänne PA, P. K., Ou I, Rybkin I, Johnson M (2019). A phase 1 clinical trial evaluating the pharmacokinetics (PK), safety, and clinical activity of MRTX849, a mutant-selective small molecule KRAS G12C inhibitor, in advanced solid tumors. In AACR-NCI-EORTC International Conference on Molecular Targets (Boston, MA). [Google Scholar]
- 65.Johung KL, Yeh N, Desai NB, Williams TM, Lautenschlaeger T, Arvold ND, Ning MS, Attia A, Lovly CM, Goldberg S, et al. (2016). Extended Survival and Prognostic Factors for Patients With ALK-Rearranged Non-Small-Cell Lung Cancer and Brain Metastasis. J. Clin. Oncol. 34, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND, Rosen EY, Richards AL, Bouvier N, Selcuklu SD, et al. (2019). Tumour lineage shapes BRCA-mediated phenotypes. Nature 571, 576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kato S, Subbiah V, Marchlik E, Elkin SK, Carter JL, and Kurzrock R (2017). RET Aberrations in Diverse Cancers: Next-Generation Sequencing of 4,871 Patients. Clin. Cancer Res. 23, 1988–1997. [DOI] [PubMed] [Google Scholar]
- 68.Khozin S, Miksad RA, Adami J, Boyd M, Brown NR, Gossai A, Kaganman I, Kuk D, Rockland JM, Pazdur R, et al. (2019). Real-world progression, treatment, and survival outcomes during rapid adoption of immunotherapy for advanced non-small cell lung cancer. Cancer 125, 4019–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D-W, Mehra R, Tan DSW, Felip E, Chow LQM, Camidge DR, Vansteenkiste J, Sharma S, De Pas T, Riely GJ, et al. (2016). Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. The Lancet Oncol. 17, 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, and Halmos B (2005). EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 352, 786–792. [DOI] [PubMed] [Google Scholar]
- 71.Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, Wasan H, Ciardiello F, Loupakis F, Hong YS, et al. (2019). Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N. Engl. J. Med. 381, 1632–1643. [DOI] [PubMed] [Google Scholar]
- 72.Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, Tada H, Kuwano H, and Mitsudomi T (2006). Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin. Cancer Res. 12, 5764–5769. [DOI] [PubMed] [Google Scholar]
- 73.Koyama T, Rhrissorrakrai K, and Parida L (2019). Analysis on GENIE reveals novel recurrent variants that affect molecular diagnosis of sizable number of cancer patients. BMC Cancer 19, 114–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lito P, Solomon M, Li L-S, Hansen R, and Rosen N (2016). Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351, 604–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu D, Offin M, Harnicar S, Li BT, and Drilon A (2018). Entrectinib: an orally available, selective tyrosine kinase inhibitor for the treatment of NTRK, ROS1, and ALK fusion-positive solid tumors. Ther. Clin. Risk Manag. 14, 1247–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Long GV, Hauschild A, Santinami M, Atkinson V, Mandalà M, Chiarion-Sileni V, Larkin J, Nyakas M, Dutriaux C, Haydon A, et al. (2017). Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N. Engl. J. Med. 377, 1813–1823. [DOI] [PubMed] [Google Scholar]
- 77.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, Garbe C, Jouary T, Hauschild A, Grob JJ, et al. (2014). Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 371, 1877–1888. [DOI] [PubMed] [Google Scholar]
- 78.Longo DL (2017). Imatinib Changed Everything. N. Engl. J. Med. 376, 982–983. [DOI] [PubMed] [Google Scholar]
- 79.Lopez JS, and Banerji U (2017). Combine and conquer: challenges for targeted therapy combinations in early phase trials. Nat. Rev. Clin. Oncol. 14, 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, Fleming M, Rezazadeh A, Mellado B, Varlamov S, et al. (2019). Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 381, 338–348. [DOI] [PubMed] [Google Scholar]
- 81.Lai GGY, Lim TH, Lim J, Liew PJR, Kwang XL, Nahar R, Aung ZW, Takano A, Lee YY, Lau DPX, et al. (2019). Clonal MET Amplification as a Determinant of Tyrosine Kinase Inhibitor Resistance in Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. J Clin Oncol. 37, 876–884. [DOI] [PubMed] [Google Scholar]
- 82.Lu H, Liu C, Velazquez R, Wang H, Dunkl LM, Kazic-Legueux M, Haberkorn A, Billy E, Manchado E, Brachmann SM, et al. (2019). SHP2 Inhibition Overcomes RTK-Mediated Pathway Reactivation in KRAS-Mutant Tumors Treated with MEK Inhibitors. Mol. Cancer Ther. 18, 1323–1334. [DOI] [PubMed] [Google Scholar]
- 83.Mai TT, and Lito P (2018). A treatment strategy for KRAS-driven tumors. Nat. Med. 24, 902–904. [DOI] [PubMed] [Google Scholar]
- 84.Mainardi S, Mulero-Sánchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, Lieftink C, Steinberg JD, de Wit N, Gonçalves-Ribeiro S, et al. (2018). SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat. Med. 24, 961–967. [DOI] [PubMed] [Google Scholar]
- 85.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues R, Robinson D, Omlin A, Tunariu N, et al. (2015). DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 373, 1697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matozaki T, Murata Y, Saito Y, Okazawa H, and Ohnishi H (2009). Protein tyrosine phosphatase SHP-2: a proto-oncogene product that promotes Ras activation. Cancer Sci 100, 1786–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCormick F (2015). KRAS as a Therapeutic Target. Clin. Cancer Res. 21, 1797–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McCusker MG, Russo A, Scilla KA, Mehra R, and Rolfo C (2019). How I treat ALK-positive non-small cell lung cancer. ESMO Open 4, e000524–e000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Swanton C, Clonal status of actionable driver events and the timing of mutational processes in cancer evolution (2015). Sci Transl Med 7 283ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Middleton PG, Mall MA, Dřevínek P, Lands LC, McKone EF, Polineni D, Ramsey BW, Taylor-Cousar JL, Tullis E, Vermeulen F, et al. (2019). Elexacaftor-Tezacaftor-Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N. Engl. J. Med. 381, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Milojkovic D, and Apperley J (2009). Mechanisms of Resistance to Imatinib and Second-Generation Tyrosine Inhibitors in Chronic Myeloid Leukemia. Clin. Cancer Res. 15, 7519–7527. [DOI] [PubMed] [Google Scholar]
- 92.Modi S, Saura C, Yamashita T, Park YH, Kim S-B, Tamura K, Andre F, Iwata H, Ito Y, Tsurutani J, et al. (2019). Trastuzumab Deruxtecan in Previously Treated HER2-Positive Breast Cancer. N. Engl. J. Med, 10.1056/NEJMoa1914510. [DOI] [Google Scholar]
- 93.Mok TS, Wu Y-L, Ahn M-J, Garassino MC, Kim HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WSME, et al. (2017). Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 376, 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mologni L, Redaelli S, Morandi A, Plaza-Menacho I, and Gambacorti-Passerini C (2013). Ponatinib is a potent inhibitor of wild-type and drug-resistant gatekeeper mutant RET kinase. Mol. Cell. Endocrinol. 377, 1–6. [DOI] [PubMed] [Google Scholar]
- 95.Moser JC, Wei G, Colonna SV, Grossmann KF, Patel S, and Hyngstrom JR (2020). Comparative-effectiveness of pembrolizumab vs. nivolumab for patients with metastatic melanoma. Acta Oncol., 1–4. [DOI] [PubMed] [Google Scholar]
- 96.Moura MM, Cavaco BM, Pinto AE, Domingues R, Santos JR, Cid MO, Bugalho MJ, and Leite V (2009). Correlation of RET somatic mutations with clinicopathological features in sporadic medullary thyroid carcinomas. Br. J. Cancer 100, 1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mulligan LM (2014). RET revisited: expanding the oncogenic portfolio. Nat. Rev. Cancer 14, 173–186. [DOI] [PubMed] [Google Scholar]
- 98.Naidoo J, Sima CS, Rodriguez K, Busby N, Nafa K, Ladanyi M, Riely GJ, Kris MG, Arcila ME, and Yu HA (2015). Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: Clinical outcomes and response to erlotinib. Cancer 121, 3212–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, Tzitzilonis C, Mordec K, Marquez A, Romero J, et al. (2018). RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat. Cell Biol. 20, 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nogova L, Sequist LV, Perez Garcia JM, Andre F, Delord J-P, Hidalgo M, Schellens JHM, Cassier PA, Camidge DR, Schuler M, et al. (2017). Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1–3 Kinase Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in Fibroblast Growth Factor Receptors: Results of a Global Phase I, Dose-Escalation and Dose-Expansion Study. J. Clin. Oncol. 35, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O’Hare T, Eide CA, and Deininger MWN (2007). Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood 110, 2242–2249. [DOI] [PubMed] [Google Scholar]
- 102.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, Adrian LT, Zhou T, Huang W-S, Xu Q, et al. (2009). AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell 16, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O’Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, Liu Y, Bartlett CH, Koehler M, Cristofanilli M, et al. (2018). Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 9, 896–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Offin M, Feldman D, Ni A, Myers ML, Lai WV, Pentsova E, Boire A, Daras M, Jordan EJ, Solit DB, et al. (2019). Frequency and outcomes of brain metastases in patients with HER2-mutant lung cancers. Cancer 125, 4380–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ortiz-Cuaran S, Scheffler M, Plenker D, Dahmen L, Scheel AH, Fernandez-Cuesta L, Meder L, Lovly CM, Persigehl T, Merkelbach-Bruse S, et al. (2016). Heterogeneous Mechanisms of Primary and Acquired Resistance to Third-Generation EGFR Inhibitors. Clin. Cancer Res. 22, 4837–4847. [DOI] [PubMed] [Google Scholar]
- 106.Ostman A, Hellberg C, and Böhmer FD (2006). Protein-tyrosine phosphatases and cancer. Nat. Rev. Cancer 6, 307–320. [DOI] [PubMed] [Google Scholar]
- 107.Ostrem JM, Peters U, Sos ML, Wells JA, and Shokat KM (2013). K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ou S-HI, Ahn JS, De Petris L, Govindan R, Yang JC-H, Hughes B, Lena H, Moro-Sibilot D, Bearz A, Ramirez SV, et al. (2016). Alectinib in Crizotinib-Refractory ALK-Rearranged Non-Small-Cell Lung Cancer: A Phase II Global Study. J. Clin. Oncol. 34, 661–668. [DOI] [PubMed] [Google Scholar]
- 109.Oxnard GR, Arcila ME, Sima CS, Riely GJ, Chmielecki J, Kris MG, Pao W, Ladanyi M, and Miller VA (2011). Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin. Cancer Res. 17, 1616–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Parikh RB, Adamson BJS, Khozin S, Galsky MD, Baxi SS, Cohen A, and Mamtani R (2019). Association Between FDA Label Restriction and Immunotherapy and Chemotherapy Use in Bladder Cancer. JAMA 322, 1209–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Patricelli MP, Janes MR, Li L-S, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, et al. (2016). Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 6, 316–329. [DOI] [PubMed] [Google Scholar]
- 112.Perez EA, Romond EH, Suman VJ, Jeong J-H, Sledge G, Geyer CE Jr., Martino S, Rastogi P, Gralow J, Swain SM, et al. (2014). Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J. Clin. Oncol. 32, 3744–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim D-W, Ou S-HI, Pérol M, Dziadziuszko R, Rosell R, et al. (2017). Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 377, 829–838. [DOI] [PubMed] [Google Scholar]
- 114.Planchard D, Besse B, Groen HJM, Souquet P-J, Quoix E, Baik CS, Barlesi F, Kim TM, Mazieres J, Novello S, et al. (2016). Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol. 17, 984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, Zhou C, Reungwetwattana T, Cheng Y, Chewaskulyong B, et al. (2020). Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 382, 41–50. [DOI] [PubMed] [Google Scholar]
- 116.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, Lichinitser M, Dummer R, Grange F, Mortier L, et al. (2015). Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 372, 30–39. [DOI] [PubMed] [Google Scholar]
- 117.Robichaux JP, Elamin YY, Tan Z, Carter BW, Zhang S, Liu S, Li S, Chen T, Poteete A, Estrada-Bernal A, et al. (2018). Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat. Med. 24, 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rodon J, Dienstmann R, Serra V, and Tabernero J (2013). Development of PI3K inhibitors: lessons learned from early clinical trials. Nat. Rev. Clin. Oncol. 10, 143–153. [DOI] [PubMed] [Google Scholar]
- 119.Rubinstein SM, and Warner JL (2018). CancerLinQ: Origins, Implementation, and Future Directions. JCO Clin. Cancer Inform. 2, 1–7. [DOI] [PubMed] [Google Scholar]
- 120.Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, Görgülü K, Dantes Z, Wörmann SM, Diakopoulos KN, et al. (2018). Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat. Med. 24, 954–960. [DOI] [PubMed] [Google Scholar]
- 121.Schapira M, Calabrese MF, Bullock AN, and Crews CM (2019). Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov. 18, 949–963. [DOI] [PubMed] [Google Scholar]
- 122.Schram AM, Chang MT, Jonsson P, and Drilon A (2017a). Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat. Rev. Clin. Oncol. 14, 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schram AM, and Hyman DM (2017). Quantifying the Benefits of Genome-Driven Oncology. Cancer Discov. 7, 552–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schram AM, Voss MH, and Hyman DM (2017b). Genome-Driven Paradigm for the Development of Selective Fibroblast Growth Factor Receptor Inhibitors. J. Clin. Oncol. 35, 131–134. [DOI] [PubMed] [Google Scholar]
- 125.Sehgal K, Patell R, Rangachari D, and Costa DB (2018). Targeting ROS1 rearrangements in non-small cell lung cancer with crizotinib and other kinase inhibitors. Transl. Cancer Res. 7, S779–S786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sequist LV, Lee JS, Han J-Y, Su W-C, Yang JC-H, Yu H, Ottesen LH, Verheijen RB, Mellemgaard A, Wessen J, et al. (2019). Abstract CT033: TATTON Phase Ib expansion cohort: Osimertinib plus savolitinib for patients (pts) with EGFR-mutant, MET-amplified NSCLC after progression on prior third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI). Cancer Research 79, CT033. [Google Scholar]
- 127.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, and Sawyers CL (2002). Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2, 117–125. [DOI] [PubMed] [Google Scholar]
- 128.Shaw AT, Gandhi L, Gadgeel S, Riely GJ, Cetnar J, West H, Camidge DR, Socinski MA, Chiappori A, Mekhail T, et al. (2016). Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol. 17, 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shaw AT, Kim D-W, Mehra R, Tan DSW, Felip E, Chow LQM, Camidge DR, Vansteenkiste J, Sharma S, De Pas T, et al. (2014). Ceritinib in ALK-rearranged non-small-cell lung cancer. N. Engl. J. Med. 370, 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shimizu T, Tolcher AW, Papadopoulos KP, Beeram M, Rasco DW, Smith LS, Gunn S, Smetzer L, Mays TA, Kaiser B, et al. (2012). The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin. Cancer Res. 18, 2316–2325. [DOI] [PubMed] [Google Scholar]
- 131.Shin HT, Choi YL, Yun JW, Kim NKD, Kim SY, Jeon HJ, Nam JY, Lee C, Ryu D, Kim SY, et al. (2017). Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Comm 8, 1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shitara K, Iwata H, Takahashi S, Tamura K, Park H, Modi S, Tsurutani J, Kadowaki S, Yamaguchi K, Iwasa S, et al. (2019). Trastuzumab deruxtecan (DS-8201a) in patients with advanced HER2-positive gastric cancer: a dose-expansion, phase 1 study. Lancet Oncol. 20, 827–836. [DOI] [PubMed] [Google Scholar]
- 133.Smyth LM, Piha-Paul SA, Won HH, Schram AM, Saura C, Loi S, Lu J, Shapiro GI, Juric D, Mayer IA, et al. (2019). Efficacy and Determinants of Response to HER Kinase Inhibition in HER2-Mutant Metastatic Breast Cancer. Cancer Discov. 10, 198–213, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spurr L, Li M, Alomran N, Zhang Q, Restrepo P, Movassagh M, Trenkov C, Tunnessen N, Apanasovich T, Crandall KA, et al. (2018). Systematic pan-cancer analysis of somatic allele frequency. Sci Rep. 8, 7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stephen AG, Esposito D, Bagni RK, and McCormick F (2014). Dragging ras back in the ring. Cancer Cell 25, 272–281. [DOI] [PubMed] [Google Scholar]
- 136.Stransky N, Cerami E, Schalm S, Kim JL, and Lengauer C (2014). The landscape of kinase fusions in cancer. Nat. Commun. 5, 4846–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, et al. (2012). RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 366, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Subbiah V, Gainor JF, Rahal R, Brubaker JD, Kim JL, Maynard M, Hu W, Cao Q, Sheets MP, Wilson D, et al. (2018a). Precision Targeted Therapy with BLU-667 for RET-Driven Cancers. Cancer Discov. 8, 836–849. [DOI] [PubMed] [Google Scholar]
- 139.Subbiah V, Velcheti V, Tuch BB, Ebata K, Busaidy NL, Cabanillas ME, Wirth LJ, Stock S, Smith S, Lauriault V, et al. (2018b). Selective RET kinase inhibition for patients with RET-altered cancers. Ann. Oncol. 29, 1869–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tamura K, Tsurutani J, Takahashi S, Iwata H, Krop IE, Redfern C, Sagara Y, Doi T, Park H, Murthy RK, et al. (2019). Trastuzumab deruxtecan (DS-8201a) in patients with advanced HER2-positive breast cancer previously treated with trastuzumab emtansine: a dose-expansion, phase 1 study. Lancet Oncol. 20, 816–826. [DOI] [PubMed] [Google Scholar]
- 141.Tao JJ, Schram AM, and Hyman DM (2018). Basket Studies: Redefining Clinical Trials in the Era of Genome-Driven Oncology. Annu. Rev. Med. 69, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Taylor MH, Gainor JF, Hu MIN, Zhu VW, Lopes G, Leboulleux S, Brose MS, Schuler MH, Bowles DW, Kim D-W, et al. (2019). Activity and tolerability of BLU-667, a highly potent and selective RET inhibitor, in patients with advanced RET-altered thyroid cancers. J. Clin. Oncol. 37, 6018–6018. [Google Scholar]
- 143.Thorpe LM, Yuzugullu H, and Zhao JJ (2015). PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 15, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.U.S. Food and Drug Administration (2018). Framework for FDA’s Real-World Evidence Program. (https://www.fda.gov/media/120060/download).
- 145.Vyse S, and Huang PH (2019). Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct. Target Ther. 4, 5–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wedam S, Fashoyin-Aje L, Bloomquist E, Tang S, Sridhara R, Goldberg KB, Theoret MR, Amiri-Kordestani L, Pazdur R, and Beaver JA (2019). FDA Approval Summary: Palbociclib for Male Patients with Metastatic Breast Cancer. Clin. Cancer Res, 10.1158/1078-0432.CCR-1119-2580. [DOI] [PubMed] [Google Scholar]
- 147.West HJ (2017). Novel Precision Medicine Trial Designs: Umbrellas and Baskets . JAMA Oncol. 3, 423–423. [DOI] [PubMed] [Google Scholar]
- 148.Wirth L, Sherman E, Drilon A, Solomon B, Robinson B, Lorch J, McCoach C, Patel J, Leboulleux S, Worden F, et al. (2019). LBA93 Registrational results of LOXO-292 in patients with RET-altered thyroid cancers. Ann. Oncol. 30. [Google Scholar]
- 149.Wirth LJ, Tahara M, Robinson B, Francis S, Brose MS, Habra MA, Newbold K, Kiyota N, Dutcus CE, Mathias E, et al. (2018). Treatment-emergent hypertension and efficacy in the phase 3 Study of (E7080) lenvatinib in differentiated cancer of the thyroid (SELECT). Cancer 124, 2365–2372. [DOI] [PubMed] [Google Scholar]
- 150.Wöhrle S, Bonny O, Beluch N, Gaulis S, Stamm C, Scheibler M, Müller M, Kinzel B, Thuery A, Brueggen J, et al. (2011). FGF receptors control vitamin D and phosphate homeostasis by mediating renal FGF-23 signaling and regulating FGF-23 expression in bone. J. Bone Miner. Res. 26, 2486–2497. [DOI] [PubMed] [Google Scholar]
- 151.Wu A-L, Feng B, Chen MZ, Kolumam G, Zavala-Solorio J, Wyatt SK, Gandham VD, Carano RAD, and Sonoda J (2013). Antibody-mediated activation of FGFR1 induces FGF23 production and hypophosphatemia. PLoS One 8, e57322–e57322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wylie AA, Schoepfer J, Jahnke W, Cowan-Jacob SW, Loo A, Furet P, Marzinzik AL, Pelle X, Donovan J, Zhu W, et al. (2017). The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature 543, 733–737. [DOI] [PubMed] [Google Scholar]
- 153.Yang J, Nie J, Ma X, Wei Y, Peng Y, and Wei X (2019). Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol. Cancer 18, 26–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M, and Riely GJ (2013). Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. 19, 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ (2007). Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 11, 217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang I, Zaorsky NG, Palmer JD, Mehra R, and Lu B (2015). Targeting brain metastases in ALK-rearranged non-small-cell lung cancer. The Lancet Oncology 16, e510–e521. [DOI] [PubMed] [Google Scholar]
- 158.Zhang S, Anjum R, Squillace R, Nadworny S, Zhou T, Keats J, Ning Y, Wardwell SD, Miller D, Song Y, et al. (2016). The Potent ALK Inhibitor Brigatinib (AP26113) Overcomes Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in Preclinical Models. Clin. Cancer Res. 22, 5527–5538. [DOI] [PubMed] [Google Scholar]
- 159.Zweig JR, and Neal JW (2018). Infiltrating the Blood-Brain Barrier in ALK-Positive Lung Cancer. J. Clin. Oncol. 36, 2677–2679. </References> [DOI] [PubMed] [Google Scholar]