Fig. 1.
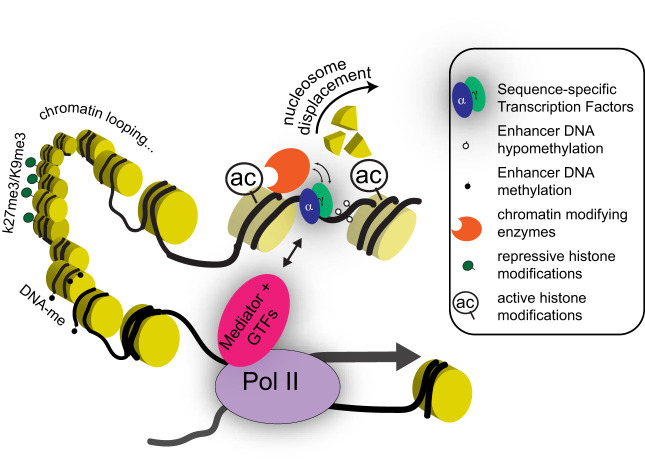
Epigenetic regulatory mechanisms discussed in this review. Gene transcription is influenced by multiple regulatory inputs coordinated by sequence-specific transcription factors (TFs) at distal cis-regulatory elements (enhancers). Chromatin accessibility is established at these genomic regions by pioneer TFs and remodeling enzymes, which together displace nucleosomes. Other TF-recruited enzymes endow neighboring nucleosomes with covalent posttranslational modifications associated with gene activity (e.g., H3K27 acetylation shown here as “ac”) or silencing (e.g., H3K27me3 and H3K9me3). In addition to carrying specific TF-binding motifs, enhancer DNA is hypomethylated at CpG dinucleotides, compared with inactive promoters or the genome background. Enhancers are thought to modulate transcription by forming three-dimensional loops that bring them close to RNA polymerase 2 (Pol II) and basal transcription machinery at target gene promoters. The protein complexes that bridge these loops contain Mediator and other general transcription factors (GTFs). Table 1 summarizes the current knowledge of these regulatory factors in the intestinal epithelium.