Extended data figure 8: Measuring spiked pseudo-contaminant contribution in downstream ML models and theoretical sensitivities of commercially available, host-based, cell-free DNA (ctDNA) assays in TCGA patients.
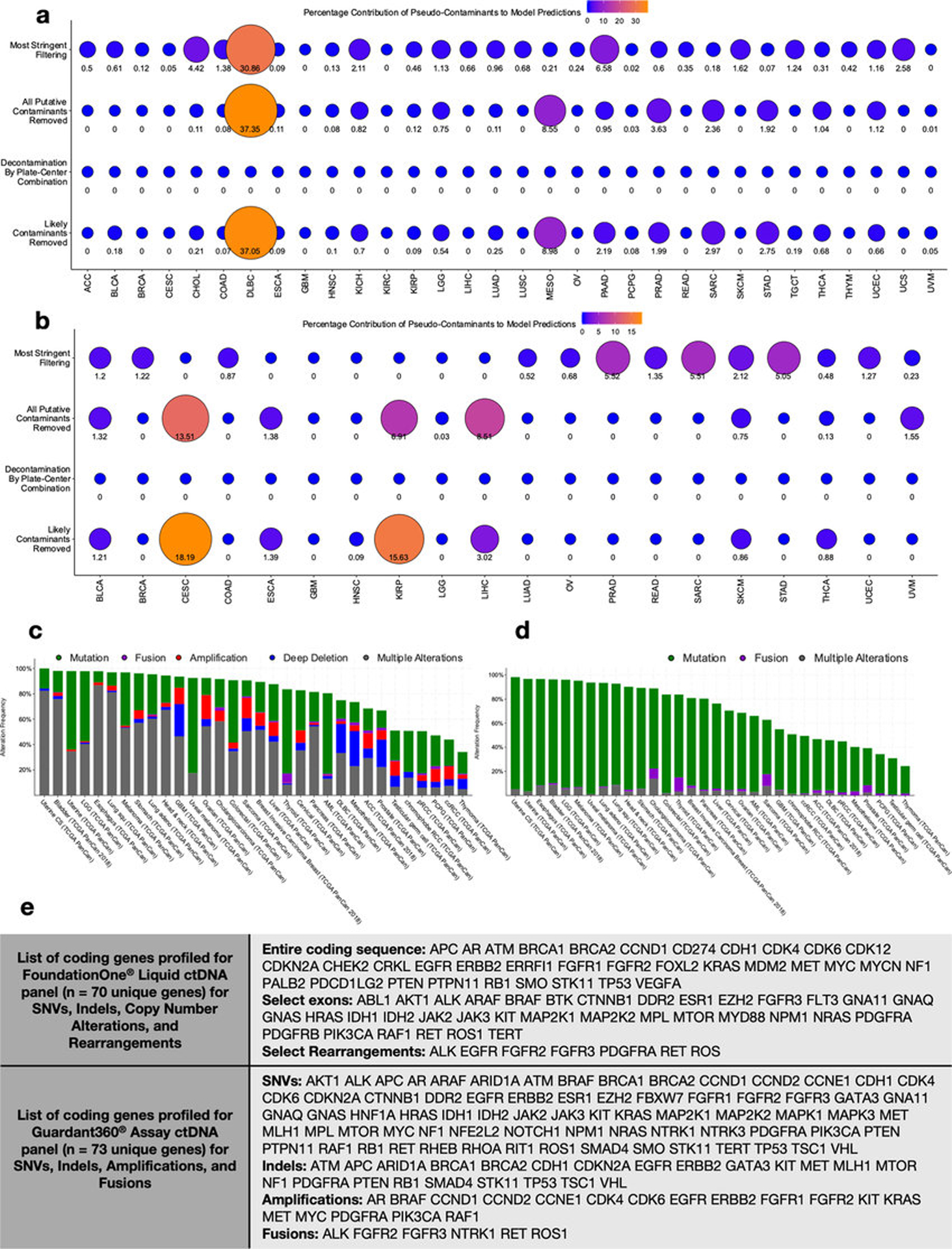
a-b, Feature importance scores were calculated for all taxa used in models trained to discriminate one-cancer-type-versus-all-others in all four decontaminated datasets (Extended Data Fig. 6b) using primary tumor microbial DNA or RNA (a), or using blood-derived mbDNA (b). These decontaminated datasets were spiked with pseudo-contaminants prior to the decontamination and normalization pipelines to evaluate their performance (Methods), and the test set performances of the models shown are given in Extended Data Figs. 6g–h and Fig. 3a, respectively. Any spiked pseudo-contaminant(s) used by a model had their feature importance score(s) divided by the sum total of all feature importance scores in that model to estimate a percentage contribution of them towards making accurate predictions; the higher the score (out of 100), the less biologically reliable the model is. Note, “0” means that no spiked pseudo-contaminants were used for making predictions by the model; none of the models generated on the “plate-center decontaminated” data included spiked pseudo-contaminants as features. c-d, Percent distribution among TCGA studies with patients having one or more genomic alterations on FoundationOne® Liquid ctDNA coding genes (c) or on Guardant360® ctDNA coding genes (d). Data are downloaded from https://www.cbioportal.org/. e, The specific list of coding genes for the FoundationOne® and Guardant360® ctDNA assays and their examined alterations (source listed in Methods).
