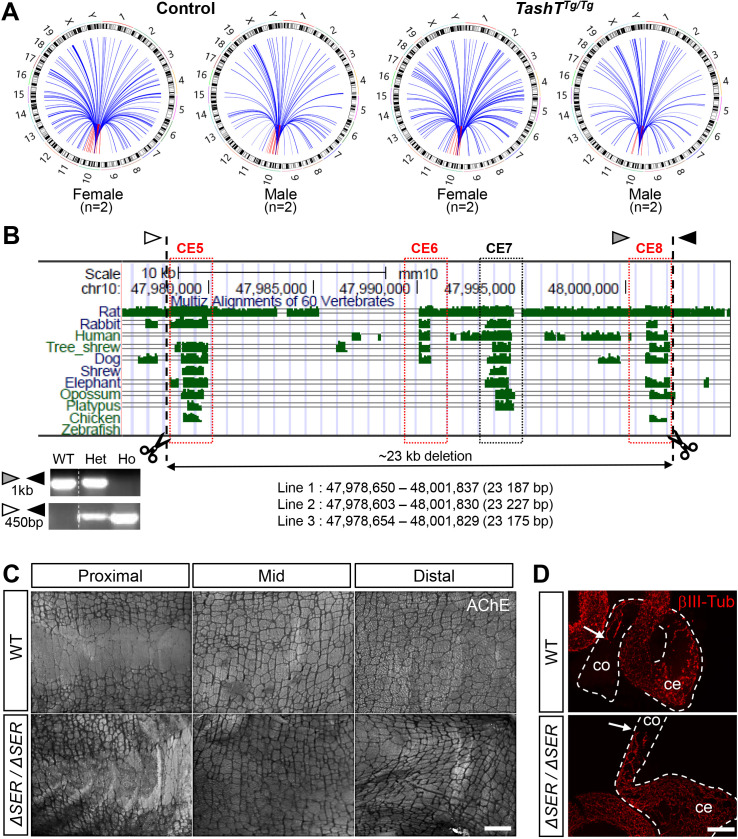
Fig 3. Ddx3y expression is not directly regulated by the Hace1-Grik2 silencer-enriched region on mouse chromosome 10.
(A) Overview of sex-stratified 4C-seq analysis of far-cis (red) and trans (blue) interactions with the Chr.10 Hace1-Grik2 silencer-enriched region in e12.5 ENCCs, which were recovered by FACS from Gata4p-GFP control and TashTTg/Tg embryos. (B) Graphical view of part of the Hace1-Grik2 intergenic region showing the relative position of five evolutionary conserved elements (CE). Those that were previously shown to possess silencer activity [29] are highlighted in red. Genomic positions of the gRNAs used for CRISPR-mediated deletion of a 23kb fragment are indicated by dashed lines. Exact deletion size is indicated for each of the three founder animals that were used to derive independent mouse lines, which were named Hace1-Grik2ΔSER/ΔSER (ΔSER/ΔSER). Arrowheads (white, grey and black) indicate the position of PCR primers used for genotyping as shown in the lower left panel; the unmodified locus gives a 1000 bp band while the CRISPR-modified locus gives a 450 bp band. WT, wild type; Het, heterozygous; Ho, homozygous. (C) Representative images of control and ΔSER/ΔSER colon tissues stained for AChE activity. Higher magnification views are shown in S9A Fig. (for FVB/N wild-type controls, n = 7 males and 6 females; for ΔSER/ΔSER mutants, n = 16 males and 15 females). (D) Representative images of βIII-Tubulin immunofluorescence labeling of e12.5 ENCCs in intestines from unsexed WT and ΔSER/ΔSER embryos (n = 12 FVB/N controls and 13 ΔSER/ΔSER mutants). Quantitative analysis of extent of ENCC colonization is shown in S9B Fig. Dashed outlines delineate the cecum (ce) and colon (co). Scale bar, 1000 μm (C); 200 μm (D).