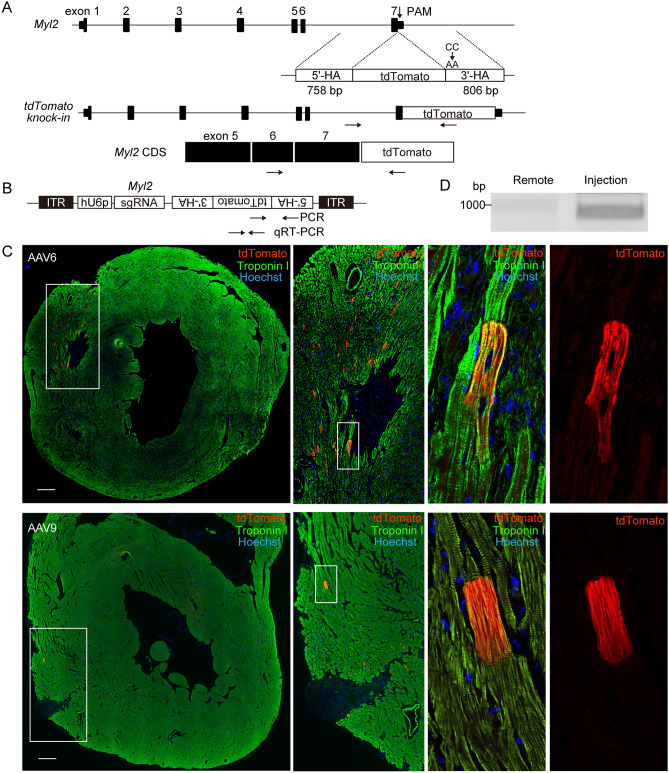
Figure 1.
Direct injection of AAV-induced precise targeted integration in adult murine heart tissues. (A) Genomic sequences of mouse Myl2 and repair template (upper) are shown, along with the expected genomic sequence after targeted integration (middle) and the 3′-terminal coding sequence (CDS) of Myl2 (lower). The repair template consisting of the coding sequence of the tdTomato fluorescent reporter protein between the 758-bp 5′-terminal and the 806-bp 3′-terminal homology arms (5′-HA and 3′-HA) corresponding to the genomic sequence of the 3′-terminus of the Myl2. PAM mutation (CC to AA) was introduced into 3′-HA, and the stop codon sequence was removed. Arrows indicate the locations of PCR primers to detect targeted integration in genomic DNA (middle) or in cDNA (lower). The forward primer to amplify genomic sequences was designed outside of 5´-HA. The primer pairs to amplify CDS were designed to span the intronic sequence. (B) The DNA sequence containing hU6 promoter (hU6p)–driven gRNA and a tdTomato repair template in the opposite direction was subcloned into AAV6 or AAV9. Arrows indicate the locations of primers to amplify the transgene by PCR and quantitative real-time PCR (qRT-PCR). (C) AAV6 (2.5 × 109 viral genome/10 μl) or AAV9 (9.2 × 1010 viral genome/10 μl) was directly injected into the free left ventricular wall. Five days after injection, heart tissues were excised and stained with anti-troponin I antibody (green). Scale bar: 300 μm. (D) Intron-spanning PCR was performed using cDNA samples obtained from AAV6-injected myocardium. cDNA obtained from remote sections that did not undergo viral injection were used as controls. Original full image of the gel is presented in Supplementary Fig. 6A.