BIOMASS YIELD 1 encodes a 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHPS) that affects sorghum biomass and grain yield through regulation of primary and secondary metabolism via the shikimate pathway.
Keywords: Biomass yield, flavonoids, metabolic profiling, metabolism, shikimate pathway, sorghum, transcriptome
Abstract
Biomass and grain yield are key agronomic traits in sorghum (Sorghum bicolor); however, the molecular mechanisms that regulate these traits are not well understood. Here, we characterized the biomass yield 1 (by1) mutant, which displays a dramatically altered phenotype that includes reduced plant height, narrow stems, erect and narrow leaves, and abnormal floral organs. Histological analysis suggested that these phenotypic defects are mainly caused by inhibited cell elongation and abnormal floral organ development. Map-based cloning revealed that BY1 encodes a 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHPS) that catalyses the first step of the shikimate pathway. BY1 was localized in chloroplasts and was ubiquitously distributed in the organs examined, particularly in the roots, stems, leaves, and panicles, which was consistent with its role in biomass production and grain yield. Transcriptome analysis and metabolic profiling revealed that BY1 was involved in primary metabolism and that it affected the biosynthesis of various secondary metabolites, especially flavonoids. Taken together, these findings demonstrate that BY1 affects biomass and grain yield in sorghum by regulating primary and secondary metabolism via the shikimate pathway. Moreover, our results provide important insights into the relationship between plant development and metabolism.
Introduction
Sorghum (Sorghum bicolor) is an annual diploid species (2n=2x=20) with a small genome (~730 Mb) and it is an agriculturally important short-day C4 plant (Paterson et al., 2009). It is one of the world’s five major food crops together with maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum), and barley (Hordeum vulgare), and is the staple food for more than 500 million people worldwide (Paterson et al., 2009; http://www.fao.org). Sorghum exhibits excellent biomass production due to its strong drought resistance, high photosynthetic efficiency, and good regeneration ability. Hence, it is a crop that is widely cultivated in arid and semi-arid areas, and it is also regarded as a bioenergy crop (Gilbert, 2009; Paterson et al., 2009; Cifuentes et al., 2014; Takaki et al., 2015; Xie and Xu, 2019). Its agricultural importance and the fact that its genome is small and has been sequenced make sorghum an excellent model organism for functional analysis of bioenergy crops (Lawrence and Walbot, 2007; Paterson et al., 2009).
Biomass and grain yield are highly complex agronomic traits that are closely related to the development of various organs and to plant metabolism. The shikimate pathway, representing a bridge between primary and secondary metabolism, is widespread in microorganisms and plants but does not exist in animals (Roberts, 1998; Tzin and Galili, 2010). Secondary metabolites accumulate in plants; in addition to playing important roles throughout growth and development, they improve resistance to invasion by pathogenic microorganisms (Hay, 1996; Kuhn et al., 2011). Flavonoids, a group of secondary metabolites including flavonols, flavones, isoflavones, and anthocyanins, are widely distributed in the flowers, fruits, and leaves of many plants. These metabolites are produced in different tissues, during specific developmental stages, and in response to various environmental factors, and they function in plant defense responses and act as messengers during plant reproduction (Taylor and Grotewold, 2005). Flavonoid biosynthesis occurs via the phenylpropanoid pathway, one of the most well-studied pathways of plant secondary metabolism, which functions downstream of the shikimate pathway (Taylor and Grotewold, 2005; Brunetti et al., 2013).
The shikimate pathway, which is required for the production of aromatic amino acids (AAAs) (Roberts et al., 1998; Yamada et al., 2008; Tzin et al., 2009; Maeda et al., 2011; Ma et al., 2012; Light and Anderson, 2013), involves a seven-step process leading to the formation of chorismate (CHR) via enzymatic reactions involving substrates of phosphoenolpyruvate (PEP) produced by the glycolysis pathway and erythritol-4-phosphate (E4-P) produced by the pentose phosphate pathway (Herrmann, 1995; Tzin and Galili, 2010). CHR, the final product of the shikimate pathway, is a common precursor of various secondary metabolites such as flavonoids, lignin, indole derivatives, and other phenolic compounds (Bentley and Haslam, 1990; Roberts et al., 1998; Ma et al., 2012; Light and Anderson, 2013). DAHPS (3-deoxy-D-arabino-heptulosonate-7-phosphate synthase), the first enzyme of the shikimate pathway, plays a central role in linking primary and specialized metabolism (Aharoni and Galili, 2011; Nazmi et al., 2014). Much is known about the function of DAHPS in various microorganisms (Maeda and Dudareva, 2012); however, it shares less than 20% sequence homology between microorganisms and plants and has obvious differences in amino acid composition and properties, suggesting that the functions may differ (Arcuri et al., 2004; Chávez-Béjar, 2008). To date, DAHPS proteins have been identified in several plant species including tobacco (Wang et al., 1991), Arabidopsis (Keith et al., 1991), potato (Zhao and Herrmann, 1992), Populus trichocarpa (Tuskan et al., 2006), grapevine (Zhang et al., 2011), tomato (Tzin et al., 2012), and rice (Pagor et al., 2015), but the specific functions and regulatory mechanisms of DAHPS in plants are not well understood.
In this study, we characterized the sorghum mutant biomass yield 1 (by1), which exhibits reduced plant height, narrow stems, narrow and erect leaves, and abnormal floral organs. Map-based cloning revealed that BY1 encodes a DAHPS. Functional analysis revealed that BY1 is constitutively expressed in sorghum and that it participates in the first step of the shikimate pathway. Transcriptome analysis and metabolic profiling suggested that BY1 influences the cell cycle and metabolite levels. Our observations suggest that BY1 is a DAHPS that influences biomass production and grain yield in sorghum by modulating primary and secondary metabolism via the shikimate pathway.
Materials and methods
Plant materials and growth conditions
In this study we used three Sorghum bicolor (Moench) parent lines, namely BTx623, the biomass yield 1 (by1) mutant, and Shangzhuang (a local broom sorghum variety in Beijing) together with two mapping populations, namely by1 × Shangzhuang and BTx623 × by1. The by1 mutant was isolated from an ethyl-methanesulfonate (EMS) mutant population developed by our group in the BTx623 background, an elite sorghum inbred line with an available genome sequence. In brief, about 4000 healthy BTx623 seeds were treated with 0.25% EMS and the mutant lines were selected from a segregating M2 population of selfed M1 plants according to various traits including tillering, panicle length, grain weight, and plant height. The by1 mutant was backcrossed with BTx623 twice to eliminate interference from other invalid mutation sites, and was then used for subsequent analyses of phenotype, mapping, expression, transcriptome, and metabonomics. An F2 mapping population was constructed by crossing by1 with Shangzhuang. All sorghum plants were grown in 2015–2019 at the Shangzhuang Experimental Station of the China Agricultural University, Beijing, China, in the summer and at another experimental field in Sanya, Hainan Province, in the winter. Plants were grown in rows at 70-cm spacing and each row contained 10 plants with 25-cm spacing. Paper bags were used to cover the panicles of the plants before flowering to prevent cross-pollination.
Phenotypic analysis
Twenty plants each of BTx623 and by1 were collected and the following agronomic traits were determined: plant height, leaf length, leaf width, internode length, internode diameter, seed-setting rate, spike length, and 1000-grain weight.
Microscopy
For histological analysis, fresh internode tissue from the middle portions of the panicle neck internodes were collected from by1 and BTx623 plants at the heading stage and fixed in 3.7% FAA solution (3.7% formaldehyde, 70% ethanol, and 5% glacial acetic acid). After incubation with shaking at 4 °C overnight, the samples were subjected to a series of dehydration and infiltration steps. The samples were embedded in paraffin, cut into 8-µm sections using a Leica RM2265 microtome, stained with 1% Safranin O and 1% Fast Green FCF, and imaged under a BX51 microscope (Olympus).
For SEM, flagleaf tissue was collected from by1 and BTx623 plants at the heading stage, cut into small pieces, and immediately fixed in 2.5% glutaraldehyde in 0.2 M phosphate buffer (pH 7.0). The samples were incubated at 4 °C overnight, washed with the same phosphate buffer, post-fixed in 1% OsO4, and dehydrated through a graded ethanol series (30–100%). The samples were then critical-point dried, coated with gold, and observed under a Hitachi S-3400N SEM. Intact, round pollen grains were considered to be fully developed, whilst those that appeared folded were counted as defective. The proportion of pollen with good integrity relative to the total number within a field of vision was calculated. The cell lengths and widths were measured using the ImageJ software.
Map-based cloning of BY1
Map-based cloning of BY1 was performed using an F2 mapping population generated by crossing the by1 mutant with the sorghum inbred line Shangzhuang. Total genomic DNA was extracted from fresh leaves using the CTAB (cetyltrimethylammonium bromide) method (Murray and Thompson, 1980), with minor modifications. Approximately 110 pairs of published SSR markers (Li et al., 2009; Yonemaru et al., 2009) with polymorphism between the parents were used for bulked segregant analysis (BSA; Michelmore et al., 1991) to identify the linkage markers. A total of 269 F2 plants were used for mapping the BY1 locus. Fine-mapping was achieved by increasing the numbers of SSR makers in 2821 F2 plants. A list of the primers used for mapping and for all other analyses is given in Supplementary Table S3 at JXB online.
Phylogenetic analysis
Using the BY1 protein sequence as a query, we searched the Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) and NCBI (https://www.ncbi.nlm.nih.gov) databases. An E-value cut-off of 0.0 was used to identify BY1-like proteins in other species. CLUSTAL W was used to align all the BY1-like protein sequences (Thompson et al., 1994). The WebLogo software was used for motif analysis (Crooks et al., 2004) and the MEGA 5.0 software was used for construction of the phylogenetic tree via the neighbor-joining method (Tamura et al., 2011).
Genetic transformation
Based on homology analysis, Loc_Os07g42960 in rice was identified as the homologous gene of sorghum by1. To generate the knock-out vector for Loc_Os07g42960, two gRNAs (Osby1-1 and Osby1-2) targeting two different sites in the first exon of Loc_Os07g42960 were designed using CRISPR-P (http://crispr.hzau.edu.cn/CRISPR2/). Osby1-1 and Osby1-2 were introduced into pYLCRISPR/Cas9-MH, a CRISPR/Cas9 binary vector. The CRISPR/Cas9 vector containing the Osby1-1 and Osby1-2 targets and an empty CRISPR/Cas9 vector were introduced into ZH11, a japonica rice variety commonly used for genetic transformation, via Agrobacterium tumefaciens EHA105-mediated transformation.
Subcellular localization of BY1
The p35S::BY1-GFP and p35S::by1-GFP vectors were constructed and used for subcellular localization analysis. The p35S::BY1-GFP vector contained the full-length cDNA sequence of BY1 fused with green fluorescent protein (GFP), and p35S::by1-GFP contained the full-length cDNA sequence of BY1 from the by1 mutant fused with GFP; both genes were driven by the 35S promoter. The vectors were introduced into maize leaf protoplasts as described by Liu et al., (2017), with minor modifications. GFP fluorescent signals were visualized under a confocal laser-scanning microscope (LSM710, Zeiss).
Enzyme activity assays of DAHPS
The enzyme activity of DAHPS was determined using a Plant 3-deoxy-D-arabinoheptulosonate-7-phosphatesynthase (DAHPS) ELISA Kit (Shenzhen Ziker Biological Co., Ltd). Young leaves from wild-type (BTx623) and by1 plants at the 6-leaf stage were ground on ice to extract total enzyme. The test samples, standard samples, and the HRP (horseradish peroxidase)-labeled test antibody were added to micropores coated with a plant anti-DAHPS antibody, incubated, and washed thoroughly. The substrate TMB (3´,3´,5´,5´-tetramethylbenzidine) was used to develop the color, after which the absorbance was measured at 450 nm using an Enzyme Standard Instrument (Power Wave XS2, BioTek), and the DAHPS activity was calculated.
RNA extraction and RT-qPCR
Total RNA was extracted and purified from various tissues of wild-type and by1 mutant plants at the booting stage, namely the roots, nodes, stems, young leaves (leaf blade), leaf sheaths, and young panicles, using Trizol reagent (Invitrogen) and an RNA-Clean Kit (Tiangen, DP412). First-strand cDNA was generated from ~2.5 μg total purified RNA by reverse-transcription using oligo(dT)18 primers (TaKaRa) and M-MLV Reverse Transcriptase (Promega, M1701). RT-qPCR was performed using a CFX96 Real-Time System (Bio-Rad), and the sorghum PP2A gene SORBI_3004G092500 was used as an internal control (Palakolanu et al., 2016). At least three biological and three technical replicates were used for all sample, and all expression data were generated using the relative quantification method (Livak and Schmittgen, 2001).
Transcriptome and metabolic profiling
Transcriptome analysis was performed by Annoroad Gene Technology (Beijing) Co., Ltd (http://www.annoroad.com/). Young leaves (~3 cm at the tip of the seventh leaf) were harvested from wild-type and by1 plants at the 6–7-leaf stage, when the sixth leaf was about 50% unfolded and about 20% of the seventh leaf had emerged. Samples from five plants were ground into a fine powder in liquid nitrogen and pooled as a single biological repeat, and three biological repeats were performed for both wild-type and by1 plants. Total RNA was extracted and purified using Trizol reagent (Invitrogen) and a RNAClean Kit (Tiangen, DP412). The libraries were constructed and sequenced using the Illumina HiSeq2000 platform. For each sample, 6 GB of RNA-seq data were obtained (~8–9× coverage of the sorghum genome). Low-quality reads (reads containing sequencing Ns>10%), short reads (Q<10 nt), and adaptor sequences were removed by filtering the raw sequences. All high-quality, clean reads were mapped to the sorghum reference genome (BTx623) using the TopHat2 software (https://ccb.jhu.edu/software/tophat/). The resulting RNA-seq data were normalized as the number of RPKM (reads per kilobase per million mapped reads) using the Cuffquant and Cuffnorm software (https://cole-trapnell-lab.github.io/cufflinks/) and used for subsequent transcriptome analysis. Significant differentially expressed genes (DEGs) between the wild-type and by1 were identified using the Cuffiinks software (Trapnell et al., 2010). Genes with at least a 2-fold change in expression and a false-discovery rate of <0.05% were defined as differentially expressed.
Metabolome analysis was performed by Wuhan Maiwei Biotechnology Co., Ltd (https://www.metware.cn/Home11.html) using a widely targeted metabolome method. All samples used for metabolome analysis were at the same stage and position as those used for the transcriptome analysis. Metabolites were quantified using a liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) system (Zhu et al., 2018). Each 100 mg sample of dry powdered tissue was extracted overnight at 4 °C with 1 ml 70% aqueous methanol solution containing 0.1 mg l–1 lidocaine. A scheduled multiple reaction monitoring method was used to quantify the metabolites (Chen et al., 2013).
Statistical analysis
The two-tailed Student’s t-tests were performed on all the data using the SPSS software.
Accession numbers
Gene sequences of by1 have been deposited in GenBank with the accession number MN832739 (by1 mutant).
Results
Characterization of the by1 mutant
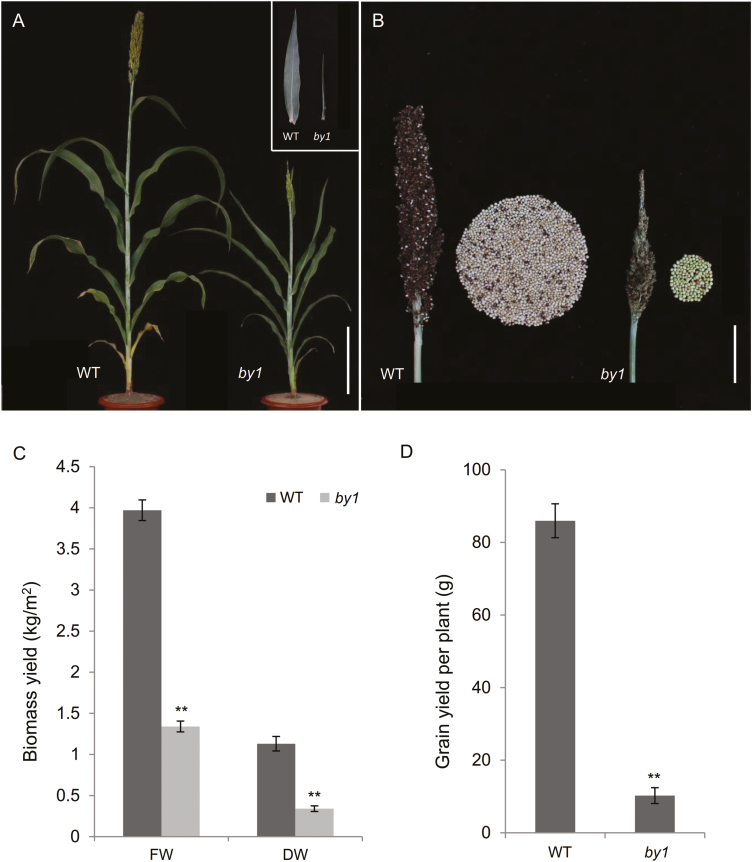
We constructed a sorghum mutant library via EMS mutagenesis of a BTx623 population (an elite sorghum inbred line with an available genome sequence) and identified the biomass yield 1 (by1) mutant. At the flowering stage, by1 plants exhibited lower height, narrower stems, and narrower leaves compared to the wild-type (WT), which resulted in a significant decrease in biomass production (Fig. 1A, C). At maturity, by1 plants exhibited a shorter panicle length, lower seed-setting rate, shorter grain length, narrower grain width, and lower 1000-grain weight than the WT, which led to a significant decrease in grain yield per plant (Fig. 1B, D, Supplementary Fig. S1E, F, Supplementary Table S1).
Fig. 1.
Biomass production and grain yield in wild-type sorghum and the by1 mutant. (A) Phenotypes of the wild-type (WT) and by1 plants at the flowering stage. Scale bar is 20 cm. (B) Phenotypes of panicles at maturity. Scale bar is 10 cm. (C) Biomass yields in terms of fresh and dry weight. (D) Gain yield (DW). Data are means (±SD), n=20. Significant differences were determined using two-tailed Student’s t-test: *P<0.05, **P<0.01.
Histological analysis
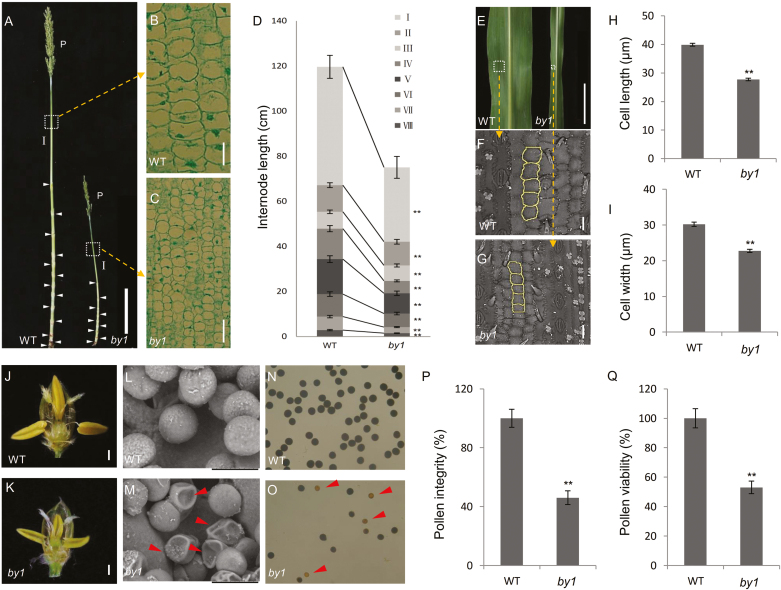
We examined the length and diameter of each internode at the flowering stage and found that all by1 internodes were shorter and smaller than those of the WT (Fig. 2A, D). To examine the reasons for the decreased plant height and thinner stems in the mutant, we performed histological analysis of the middle portions of the panicle neck internodes. Although the internode cells of both the WT and by1 were well developed, the cell volume was significantly reduced in the mutant (Fig. 2B, C, Supplementary Fig. S1A–D). Compared to the WT, the leaf length and width were significantly reduced in by1. We performed SEM and paraffin sectioning to examine leaf-blade and leaf-vein cells, respectively, and found that both were well developed in both the genotypes; however, the cell lengths and widths were significantly reduced in by1, resulting in a smaller cell volume compared to the WT (Fig. 2E–I; Supplementary Fig. S2).
Fig. 2.
Histological analysis of wild-type sorghum and the by1 mutant. (A) Stems of the wild-type (WT) and by1. White arrowheads indicate nodes. Scale bar is 20 cm. (B, C) Longitudinal sections of the panicle neck internodes of the WT and by1. Scale bars are 50 µm. (D) Comparison of internode lengths between the WT and by1. (E) Leaves of the WT and by1 (scale bar is 10 cm), and (F, G) corresponding SEM images, with cell morphologies highlighted (scale bars are 30 µm). (H) Leaf cell lengths and (I) cell widths in the WT and by1. (J, K) Images of fertile florets in the WT and by1. Scale bars are 1 mm. (L, M) Images of pollen grains of the WT and by1. Scale bars are 50 µm. (N, O) Staining for pollen viability in the WT and by1. (P) Pollen integrity, as determined from images in (L, M). (Q) Pollen viability, as determined from images in (N, O). Data are means (±SD), n=10, except n=20 in (D). Significant differences were determined using two-tailed Student’s t-test: *P<0.05, **P<0.01.
In addition to the differences in plant height and in the leaves, the grain yield was significantly reduced in by1. Comparing the young panicles of WT and by1 plants before flowering, we found severe developmental defects in the mutant, especially at the tips of panicles (Supplementary Fig. S3). We also compared the phenotypes of fertile florets at the flowering stage, and found that all those of the WT were well developed and their anthers were golden-yellow and plump, whereas those of by1 showed various developmental defects and their anthers were light-yellow, thin, and small (Fig. 2J, K; Supplementary Fig. S4). Under SEM, the pollen grains of by1 were found to be smaller than those of the WT, and ~50% of them appeared crumpled (Fig. 2L, M, P). When stained with 1% I2-KI solution, almost all the WT pollen appeared dark brown, indicating viability, whereas only 50% of by1 pollen was stained (Fig. 2N, O, Q). Overall, these results suggested that the phenotypic changes in the by1 mutant were the result of reduced cell elongation and abnormal development of the flower organs.
Map-based cloning of BY1
To determine the genetic characteristics associated with the altered phenotypes, we backcrossed by1 plants with the WT, BTx623. All F1 plants showed WT phenotypes, whilst ~25% of plants in the F2 population exhibited by1 phenotypes (83 by1 plants and 241 WT plants; χ2c =0.037, χ20.05,1=3.84). This segregation was very close to the hypothesized ratio of 3:1, indicating that the altered phenotypes of by1 were caused by a single, recessive Mendelian gene.
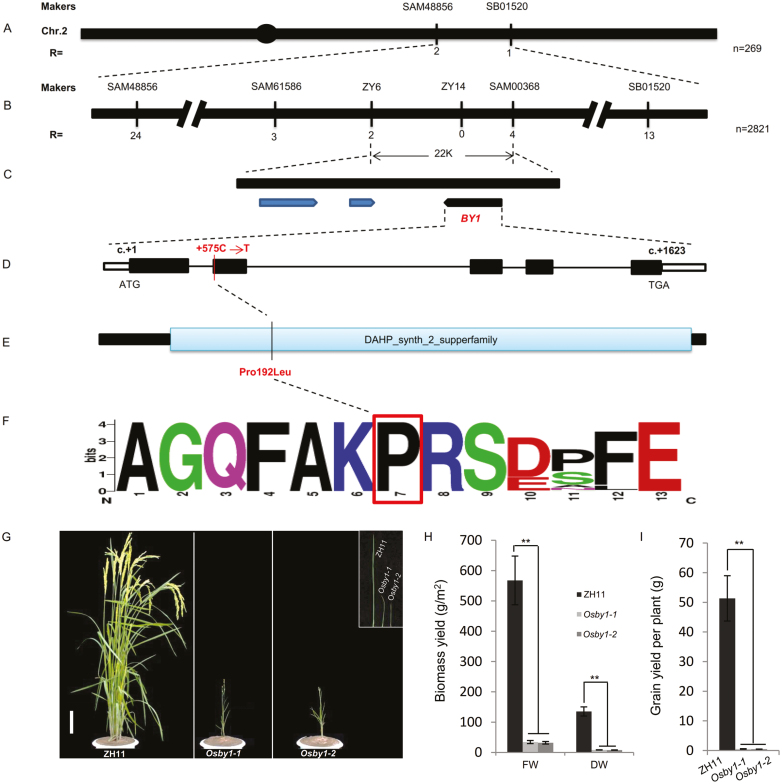
To clone BY1, we generated another F2 population derived from a cross between by1 and Shangzhuang, a local broom sorghum variety. Using map-based cloning, we roughly mapped the BY1 gene to the end of the long arm of chromosome 2, in a region between the simple-sequence repeat (SSR) markers SAM48855 and SB01520. To fine-map the BY1 locus, we then used another 2821 additional individuals from the same F2 population for recombinant screening, and the BY1 gene was further delimited to a 22-kb region between the SSR markers ZY6 and ZY21. Three predicted ORFs were located in this region according to the sorghum genome sequence database (https://phytozome.jgi.doe.gov/pz/portal.html) (Fig. 3A–C). Sequencing of the mapped genomic regions in by1 and the WT revealed only one single-nucleotide polymorphism (SNP). A transformation from C to T at position +575 (from the ‘A’ in the start codon of BY1) in exon 2 of Sobic.002G379600 caused a Pro192Leu amino acid substitution in the mutant (Fig. 3D, E), indicating that this was the the potential candidate gene for BY1.
Fig. 3.
Map-based cloning of sorghum BY1. (A) Rough and (B) fine-mapping of BY1, where n is the total number of plants used for recombinant screening and mapping, and R is the number of recombinant individuals identified by screening recombinants between flanking markers. (C) Genes in the fine-mapping region of BY1. (D) The structure of BY1 and the by1 mutation site. The unfilled boxes at the left and right ends represent the 5´-UTR and 3´-UTR, respectively. The black boxes represent exons and the lines between them represent introns. The red line and text at the second exon indicate the position and form of base mutation in the by1 mutant. (E) The amino acid structure of BY1 and the mutation site in by1. The blue box represents the functional domain region, and indicates the position and form of the amino acids in the by1 mutant. (F) Conservation analysis of the amino acid substitution region and frequency in homologous plant genes. The red box indicates the position of the amino acid substitution in by1. (G–I) The japonica rice cultivar ZH11 was gene-edited using CRISPR/Cas9. Osby1-1 is a line containing target 1 and Osby1-2 is a line containing target 2. (G) Phenotypes of the lines. The scale bar is 5 cm. (H) Biomass yield and (I) grain yield of the lines (both DW). Data are means (±SD), n=10. Significant differences compared with the ZH11 wild-type were determined using two-tailed Student’s t-test: **P<0.01.
To verify that Sobic.002G379600 was BY1, we compared its sequence in 164 lines of the sorghum germplasm collected worldwide, and found that they all showed the same genotype as the WT at the site of the by1 mutation (Supplementary Data S1). We randomly selected and sequenced 20 recessive plants from the by1 × Shangzhuang F2 population And found that a;; of them contained the same mutation site as by1 (Supplementary Fig. S5). These results suggested that the by1 mutation was induced by EMS and that it may not exist in natural sorghum accessions. Furthermore, phylogenetic analysis showed that BY1 is widely conserved in plants and shares the closest relationships with genes from other Poaceae, including Panicum virgatum, Pa. hallii, Setaria viridis, Se. italic, Zea mays, Oropetium thomaeum, O. sativa, Brachypodium stacei, and B. distachyon (Supplementary Fig. S6). Importantly, multiple-sequence alignment and motif analysis revealed that the 192Pro amino acid is conserved in higher plants (Fig. 3F). These results implied that the Pro192Leu amino acid substitution in by1 may be responsible for its growth defects.
Since it is difficult to perform genetic transformation in sorghum and given that BY1 is highly conserved between sorghum and rice, we used the japonica rice variety ZH11 (commonly used in genetic studies) as the recipient material for transformation. We generated a knock-out construct using the binary vector pYLCRISPR/Cas9-MH containing two different gRNAs, named OsBY1-1 and OsBY1-2. We introduced this construct into ZH11 to knock out Loc_Os07g42960, the homologous gene of sorghum by1 in rice, and obtained 29 successfully edited plants. These plants exhibited seven different genotypes, as revealed by sequencing. As expected, all of the plants showed dwarfing, narrow leaves, poor spike development, and possessed significantly reduced biomass and grain yield compared to the WT, making them highly similar to the phenotypes of the sorghum by1 mutant (Fig. 3G–I, Supplementary Fig. S7).
These results suggested that Sobic.002G379600 is BY1, which regulates biomass and grain yield in sorghum, and that the function of BY1 is conserved between sorghum and rice.
Transcriptional and functional characterization of BY1
To obtain the full-length BY1 sequence, we used the RACE technique. The cDNA of WT BY1 is 2039 bp long, with a 77-bp 5′-untranslated region, a 1623-bp ORF, and a 339-bp 3′-untranslated region. The gene contains five exons and four introns and encodes a 540 amino-acid protein (Supplementary Fig. S8).
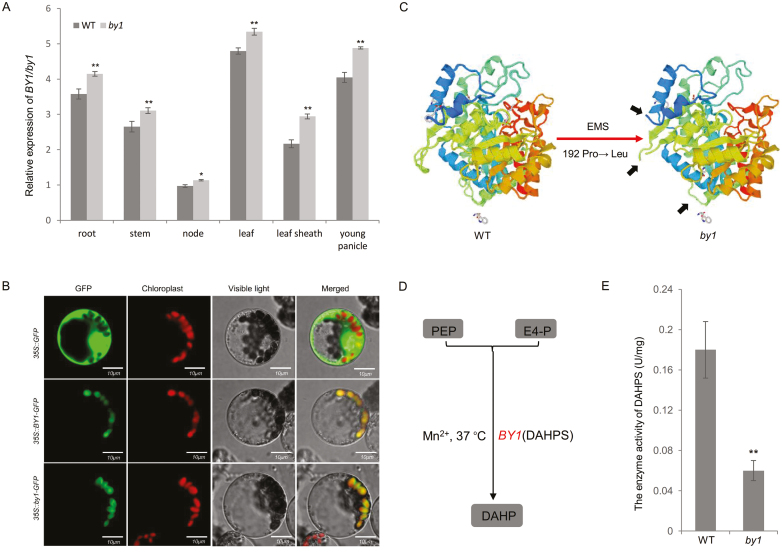
To investigate the expression patterns of BY1, we performed a RT-qPCR analysis and found that it was constitutively expressed in the roots, stems, nodes, leaves, leaf sheaths, and young panicles. Compared to the WT, BY1 was slightly up-regulated in all these tissues in the by1 mutant (Fig. 4A). We examined subcellular localization by fusing the coding sequence of BY1 in the WT and by1 to GFP to generate the fusion constructs p35S::BY1-GFP and p35S::by1-GFP, respectively, and transferred these into maize protoplasts. The BY1 fusion proteins were specifically localized to chloroplasts, and the by1 mutation did not affect this (Fig. 4B).
Fig. 4.
Transcriptional and functional characterization of BY1. (A) Relative expression of BY1 in different tissues of the sorghum wild-type (WT) and the by1 mutant. (B) Subcellular localization of the BY1 and by1 proteins in maize protoplasts. 35S::GFP is the control (top row), 35S::BY1-GFP fused with the full-length sequence of BY1 is the WT gene fusion (middle row), and 35S::by1-GFP fused with the full-length sequence of by1 is the mutant gene fusion (bottom row). Scale bars are 10 μm. (C) Comparison of the tertiary structure of the BY1 protein in the WT with by1 in the mutant. The structures were predicted using SWISS-MODEL (https://swissmodel.expasy.org/). Black arrows indicate the obvious structural alterations between the WT and by1 due to the conversion of the 192nd amino acid from Pro to Leu. (D) Reaction scheme for 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHPS, BY1). PEP, phosphoenolpyruvate; E4-P, erythrose-4-phospate. (E) Enzyme activity of DAHPS (BY1) in the WT and the by1 mutant. Data are means (±SD), n=3. Significant differences were determined using two-tailed Student’s t-test: *P<0.05, **P<0.01.
Since the mutation site occurred in the conservative domain of BY1 and the physical and chemical properties of Pro and Leu are quite different, we hypothesized that the mutation might result in a structural change in the BY1 protein. To test this, we used SWISS-MODEL (https://swissmodel.expasy.org/) to examine the tertiary structure and found that it was indeed significantly altered when the 192nd amino acid was changed from Pro to Leu (Fig. 4C; Supplementary Fig. S15).
Protein BLAST analysis of the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/) and Pfam analysis showed that BY1 encodes a DAHPS, which catalyses the formation of 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) from phosphoenolpyruvate (PEP) and erythrose-4-phospate (E4-P) (Fig. 4D). To determine whether the mutation led to loss of function, we assayed the enzyme activity of BY1 with PEP and E4-P. Compared to the WT, the enzyme activity of the by1 mutant was significantly reduced, implying that BY1 lost its normal function as a result of the mutation (Fig. 4E).
Transcriptome analysis and metabolic profiling
To elucidate the molecular mechanism underlying how BY1 influences development in sorghum, we performed transcriptome analysis and metabolic profiling using the leaves of WT and by1 plants at the 6–7-leaf stage, when the phenotypes of the plants were markedly different. We examined three biological replicates, with each sample consisting of five individual leaves of different plants at the same age and position.
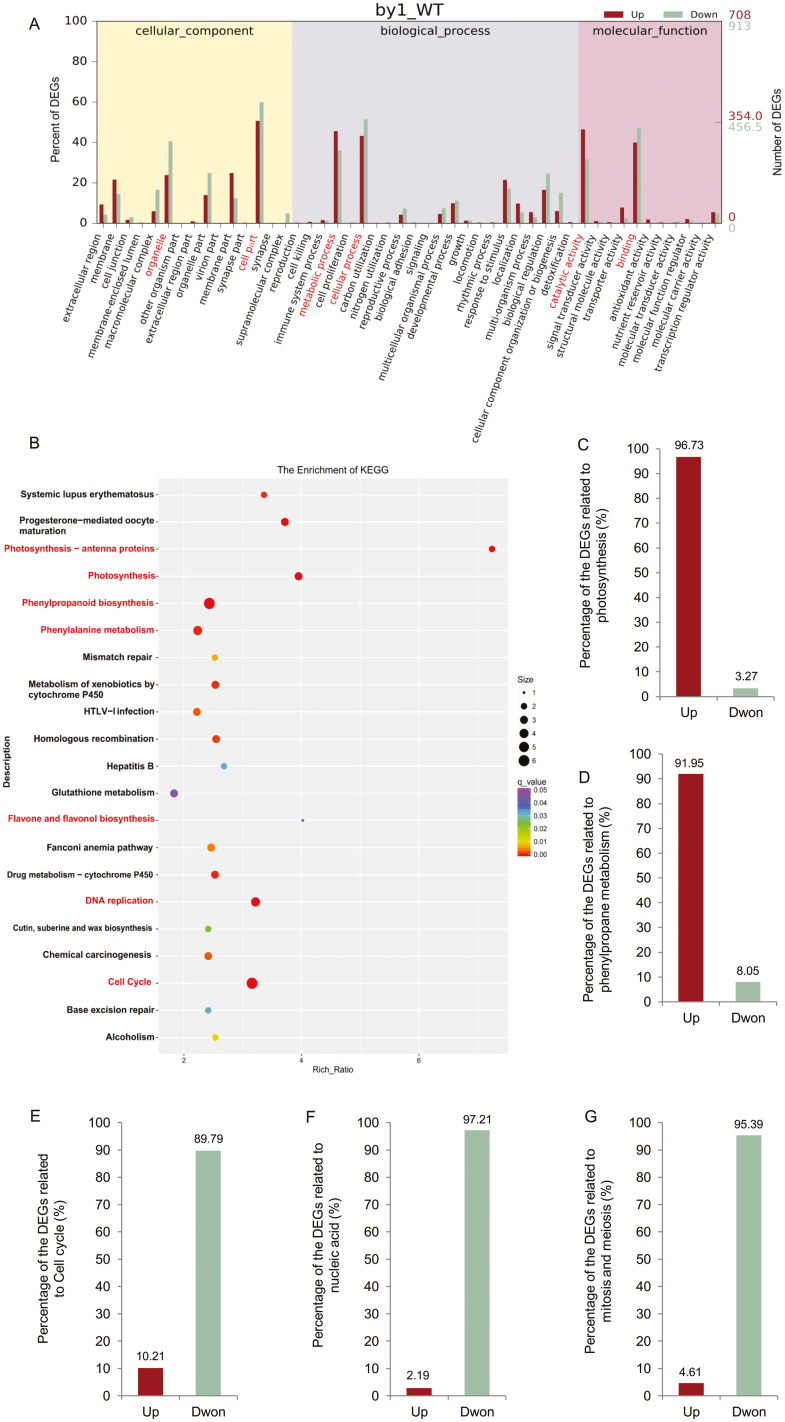
We defined significantly differentially expressed genes (DEGs) as those with a fold-change ≥2 or ≤0.5, and q<0.05. Compared to the WT, a total of 1785 DEGs were detected in all three biological replicates in by1, of which 851 were up-regulated and 934 were down-regulated (Supplementary Fig. S9A, B, Supplementary Data S2). Using the hierarchical clustering method, we clustered the DEGs into six subclasses based on their expression levels (Supplementary Fig. S9C). We then performed gene ontology (GO) analysis and divided the DEGs into the categories ‘biological process’ (BP), ‘molecular function’ (MF), and ‘cellular component’ (CC). The DEGs were enriched in many different GO terms, especially ‘cell part’ and ‘organelle’ in the CC category, ‘metabolic process’ and ‘cellular process’ in the BP category, and ‘catalytic activity’ and ‘binding’ in the MF category (Fig. 5A).
Fig. 5.
Transcriptome analysis of sorghum BY1. (A) Gene ontology (GO) analysis of differentially expressed genes (DEGs). The numbers of up- and down-regulated DEGs in each functional pathway are indicated on the right axis, and their percentages are indicated on the left axis. (B) q-values of individual groups of enriched KEGG pathways. Each point represents the degree of enrichment of the KEGG category according to the color scale, whilst the number of DEGs is indicated by the size of the point. (C–G) Percentages of up- and down-regulated DEGs related to (C) photosynthesis, (D) phenylpropane metabolism, (E) the cell cycle, (F) nucleic acids, and (G) mitosis and meiosis.
GO enrichment and KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis demonstrated that BY1 influenced various biological pathways, including photosynthesis, the cell cycle and division, and primary and secondary metabolism (Fig. 5B, Supplementary Fig. S9D, E). We further explored the expression patterns of DEGs related to photosynthesis, the cell cycle, phenylpropane metabolism, nucleic acid, and mitosis and meiosis. Interestingly, almost all DEGs related to energy biosynthesis pathways, such as photosynthesis and phenylpropane metabolism, were significantly up-regulated in by1 compared with the WT (Fig. 5C, D, Supplementary Fig. S10A, B). In contrast, almost all the DEGs involved in energy consumption pathways, such as the cell cycle, nucleic acid processes, and mitosis and meiosis, were down-regulated in the mutant (Fig. 5E–G, Supplementary Figs S10C, S11A, B).
As BY1 catalyses the formation of DAHP from PEP and E4-P and participates in the first step of the shikimate pathway, we investigated the expression patterns of genes involved in this pathway, together with those involved in the pentose phosphate pathway and glycolysis, and found that all were significantly up-regulated in by1compared the WT (Supplementary Fig. S12A–C). In addition, we examined the expression patterns of genes downstream in the general phenylpropanoid pathway and found that that they were also up-regulated in the mutant (Supplementary Fig. S12D). Finally, to validate the reliability of the transcriptome data, we randomly selected 10 up-regulated and 10 down-regulated DEGs and carried out RT-qPCR analysis. As expected, the relative expression levels were generally consistent with the mRNA-seq data (Supplementary Fig. S11C, D), suggesting that the data were of high quality.
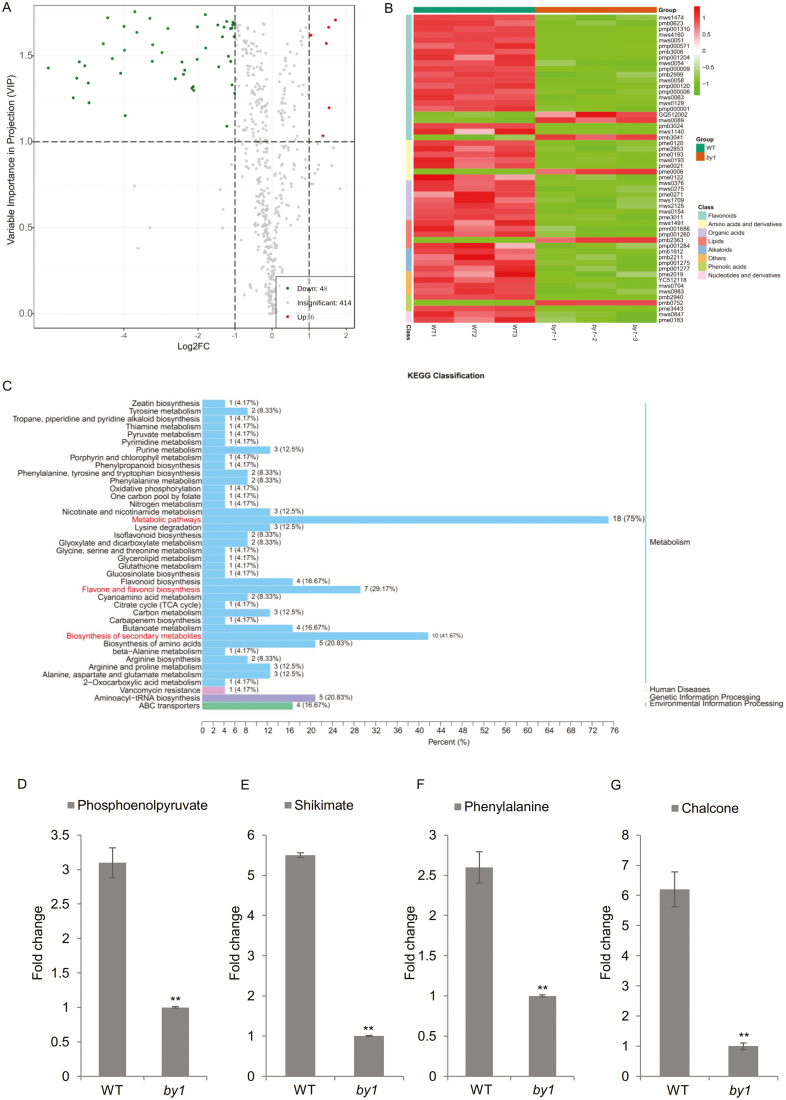
To further assess the influence of BY1 on metabolites in sorghum, we performed a widely targeted metabolomics analysis using a UPLC-MS/MS detection platform. We detected 414 differential metabolites in leaves of the WT compared with by1, (Fig. 6A) including flavonoids, organic acids, amino acids and their derivatives, phenolic acids, lipids, alkaloids, and nucleotides and their derivatives. Principal component analysis (PCA) revealed an obvious separation between the WT and by1 samples, indicating that they differed significantly (Supplementary Fig. S13A).
Fig. 6.
Metabolic profiling analysis of sorghum wild-type BY1 and the by1 mutant. (A) Volcano plot of the different metabolites. For each metabolite, the variable importance in projection (VIP) value is plotted against the logarithm of the quantitative difference multiple of the metabolite in two samples: the larger the absolute value, the greater the difference multiple of the content of the metabolite. The larger the VIP value, the more significant the difference in the metabolite levels. Metabolites down-regulated in by1 relative to the wild-type (WT) are shown in green, metabolites up-regulated are shown in red, and those with no significant difference are shown in gray. (B) Clustering heatmap of metabolites with significantly different contents (MSDCs) in by1 compared with the WT. The metabolite classes are grouped according to the color scale and their contents are indicated by the red–green scale. WT lines are to the left and by1 lines are to the right. (C) KEGG classification of MSDCs. The number of the MSDCs annotated to each pathway is shown together with its percentage of the total number of MSDCs. (D–G) Fold-changes of (D) phosphoenolpyruvate, (E) shikimate, (F) phenylalanine, and (G) chalcone levels between the WT and by1. Data are means (±SD), n=3. Significant differences were determined using two-tailed Student’s t-test: **P<0.01.
Based on the criteria of a fold-change of ≥2 or ≤0.5, we identified 54 metabolites with significantly different levels between the genotypes, with six having higher levels and 48 having lower levels in by1 (Fig. 6A; Supplementary Table S2); we termed these 54 metabolites as ‘metabolites with significantly different contents’ (MSDCs). The MSDCs were clustered into six subclasses based on the changes in their levels (Supplementary Fig. S13B). To observe these changes more clearly, we normalized the levels, divided them into different classes, and drew a heatmap using the R software. The levels of most MSDCs were significantly reduced in by1 compared with the WT. The largest class was flavonoids (Fig. 6B). KEGG analysis revealed that the MSDCs were enriched in various pathways, with the most highly enriched being the metabolic pathways in the biosynthesis of secondary metabolites, and in flavone and flavonol biosynthesis (Fig. 6C, Supplementary Fig. S13C).
Since BY1 is a functional enzyme that participates in the shikimate pathway, which functions downstream of the pentose phosphate pathway and glycolysis, we investigated the levels of metabolites in these primary metabolic pathways. Compared to the WT, the content of shikimic acid (the product of the shikimate pathway) was reduced by ~5.5-fold in by1, and the content of PEP (the intermediate product of glycolysis) was reduced by ~3.1-fold (Fig. 6D, E). In addition, the levels of many metabolites in pathways downstream of the shikimate pathway were also significantly reduced in by1. For example, the content of phenylalanine in the AAA pathway was ~2.6-fold lower and the content of chalcone in the phenylpropane pathway was ~6.2-fold lower (Fig. 6F, G). Finally, most of the MSDCs were secondary metabolites involved in phenylpropane metabolism, which also occurs downstream of the shikimate pathway. Of these metabolites, all four alkaloids and 19 of the 22 flavonoids accumulated to lower levels in by1 than in the WT (Supplementary Table S2). These results indicated that the mutation in by1 significantly influenced the homeostasis of primary and secondary metabolites in sorghum.
Discussion
Sorghum is one of the world’s most important crops, providing staple food for more than 500 million people, as well as serving as a model energy plant with high biological yield (Anami et al., 2015). Biomass and grain yield are thus key agronomic traits in sorghum; however, the molecular mechanisms that regulate them are not well understood. Here, we isolated the by1 mutant, cloned BY1, and examined its function. BY1, a constitutively expressed gene, encodes a 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHPS) that is localized to chloroplasts. Transcriptome analysis and metabolic profiling shed light on how BY1 regulates sorghum biomass and grain yield, and provided important insights into the relationship between plant development and metabolism.
Abnormal cell cycle and floral organ development are responsible for the phenotype of the by1 mutant
The biomass of by1 was reduced compared to the wild-type (WT) for several reasons, including morphological abnormalities, reduced plant height, thin stems, and narrow leaves (Fig. 1A, C). In addition, poorer development of the top of the panicle, shorter panicle length, and lower seed-setting rate in by1 led to decreased grain yield per plant (Fig. 1B, D, Supplementary Fig. S3). At the microscopic level, paraffin-sectioning and SEM analysis revealed that the decreased plant height, thinner stems, and narrower leaves in by1 were due to inhibited cell elongation (Fig. 2A–I, Supplementary Figs S1A–D, S2), which ultimately resulted in the reduced biomass. The decreased grain yield in by1 was due to poor anther development, incomplete pollen development, and decreased pollen quality and quantity (Fig. 2J–Q). At the gene-expression level, transcriptome analysis indicated the presence of many DEGs related to the cell cycle, pollen development, nucleic acids, and mitosis and meiosis, most of which were down-regulated in by1 relative to the WT (Fig. 5A, B, E, Supplementary Figs S9D, S10C, S11A, B); these findings were all consistent with the phenotypes of the mutant.
A single-nucleotide mutation leads to BY1 loss of function
We cloned sorghum BY1 using map-based cloning and based on Pfam analysis, determined that it encodes a member of the DAHP_synth_2_supperfamily (Fig. 3A–E). Homology analysis indicated that the DAHP_synth_2_supperfamily is widespread in higher plants (Supplementary Fig. S6), whilst motif analysis and multiple sequence alignment demonstrated that the amino acid mutation site in by1 is located in a region that is highly conserved among higher plants (Fig. 3F). Comparison of 164 sorghum germplasm strains and 20 randomly selected recessive plants from an F2 population suggested that the mutation in by1 may not exist in natural sorghum accessions, and that the mutation site was likely to play an important role in maintaining the normal function of BY1 (Supplementary Data S1, Supplementary Fig. S5).
Analyses of gene expression and subcellular localization indicated that BY1 was constitutively expressed and that BY1 was specifically localized to chloroplasts, with its subcellular location not being affected by the by1 mutation (Fig. 4A, B). The conversion of the 192nd amino acid from Pro to Leu strongly altered the tertiary structure of the BY1 protein (Fig. 4C, Supplementary Fig. S15), which abolished its enzymatic activity (Fig. 4D, E). Finally, the phenotypes of all the independently gene-edited rice plants that we examined were highly similar to those of the sorghum by1 mutant (Fig. 3G–I, Supplementary Fig. S7). Taken together, results demonstrated that Sobic.002G379600 was BY1, that this gene influenced biomass and grain yield in sorghum, and that a single-nucleotide mutation was responsible for its loss of function.
BY1 participates in the shikimate pathway and influences primary and secondary metabolism
The shikimate pathway only exists in microorganisms and plants (Bickel and Schultz, 1979; Maeda and Dudareva, 2012). This pathway initiates from PEP and E-4-P and ends with chorismate (CHR), and it involves seven reactions catalysed by six different enzymes (Supplementary Fig. S14A). Phenylalanine and tryptophan are essential amino acids for animals that must be obtained from food, and tyrosine is synthesized from phenylalanine. In plants, the shikimate pathway is a more widespread metabolic pathway than nitrogen fixation and photosynthesis (Jensen, 2006; Maeda and Dudareva, 2012), as it is required to generate aromatic amino acids (AAAs), which are crucial for both plants and animals. The shikimate pathway plays an important role in regulating primary and secondary metabolism (Zabalza et al., 2017).
Our results provide several lines of evidence that BY1 is involved in the shikimate pathway and influences primary and secondary metabolism (Supplementary Fig. S14). First, we have demonstrated that BY1 encodes a DAHPS protein, and that the mutation in by1 leads to loss of function (Figs 3, 4C, D) and the induction of genes involved in the shikimate pathway (Supplementary Figs S12C, S14B). Second, transcriptome analysis indicated that most genes related to photosynthesis, glycolysis, and the pentose phosphate pathway upstream of the shikimate pathway, and the phenylpropane metabolism pathway downstream of the shikimate pathway, were significantly up-regulated in the by1 mutant (Fig. 5C, D, Supplementary Figs S10A, B, S12A, B, D). Third, metabolic profiling revealed that the contents of several primary metabolites were reduced in by1 compared with the WT. For example, levels of shikimate, PEP, and phenylalanine in the mutant were reduced by ~5.5-fold, ~3.1-fold, and ~2.6-fold, respectively (Fig. 6D–F, Supplementary Fig. S14A), and the levels of most flavonoids (secondary metabolites) were also significantly reduced in by1 (Fig. 6B–G, Supplementary Fig. S14A, Supplementary Table S2).
Based on our comprehensive analyses of the changes in metabolite contents in the shikimate pathway and its upstream and downstream pathways, together with the changes in the expression of genes related to these pathways, we propose a metabolic map that highlights the important regulatory role played by BY1 (Fig. S14). , which
In summary, our results indicated that BY1 participates in the shikimate pathway and regulates the flow of primary carbon metabolites into the biosynthesis of CHR, which then generates AAAs and secondary metabolites.
A possible feedback regulation mechanism in sorghum primary and secondary metabolism
The results of transcriptome analysis and metabolic profiling indicated that genes involved in glycolysis, and the pentose phosphate, shikimate and phenylpropanoid pathways were up-regulated in the by1 mutant. However, the levels of metabolites in these pathways were reduced (PEP, shikimate, chalcone, and flavonoids). These results suggest that a feedback mechanism might exist in sorghum to regulate primary and secondary metabolism. In the WT, the functioning of BY1 was required for maintenance of the homeostasis in gene expression and the flux of metabolites, and hence for normal growth and development. However, in the by1 mutant, the abnormal functioning of BY1 and the compromised shikimate pathway may have triggered the metabolic adjustments that were observed in primary and secondary metabolism, resulting in the decreased contents of the associated metabolites. The decreased metabolic levels, or changes in associated redox states, may have been sensed by as yet unknown mechanisms to regulate gene expression in order to restore the carbon flux through the pathways. Our results agree with previous findings that reduced levels of AAAs or downstream products induce gene expression in the shikimate pathway (Maeda and Dudareva, 2012).
A model for the role of BY1 in regulating plant growth and development in sorghum
More than 90% of the total metabolic energy in bacteria is used to synthesize proteins (Herrmann and Weaver, 1999), and the shikimate pathway is used almost exclusively to synthesize AAAs, which are further used to synthesize proteins (Garbe et al.,1991; Herrmann and Weaver, 1999).In contrast, in higher plants most AAAs synthesized by the shikimate pathway are used as precursors of secondary metabolites (Maeda and Dudareva, 2012). Approximately 20% of the carbon fixed in green plants is estimated to pass through the shikimate pathway. Every year, ~7×1015 kg of carbon enters to this pathway worldwide, most of which eventually forms AAAs, flavonoids, vitamins, lignin, phenols, and other secondary metabolites that regulate plant growth and development (Rippert et al., 2004).
The amount and types of secondary metabolites synthesized by the shikimate pathway vary greatly from species to species (Campbell et al., 2004). Flavonoids are secondary metabolites that are widely distributed in plants. In addition to functioning as antioxidants that help plants cope with abiotic stress, they regulate plant growth and development by participating in the transmission of hormonal signals, promoting pollen tube germination, and sensing microbial signals (Taylor and Grotewold, 2005; Kuhn et al., 2011; Brunetti et al., 2013).
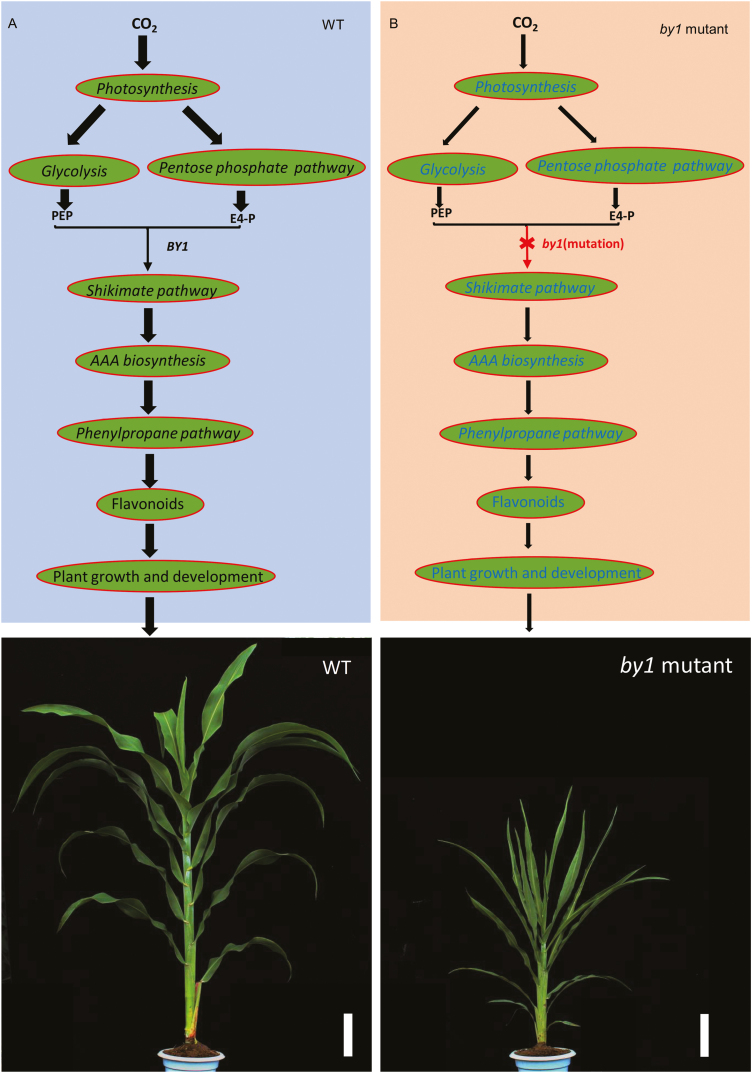
Based on the findings of our study, we propose a model to describe how BY1 regulates growth and development in sorghum (Fig. 7). According to this model, a portion of the carbon fixed by photosynthesis flows into the shikimate pathway through glycolysis and the pentose phosphate pathway and is used to form AAAs. These AAAs are then used to synthesize various secondary metabolites (especially flavonoids) via phenylpropane metabolism to regulate plant growth and development (Fig. 7A). In the BY1 loss-of-function mutant, the shikimate pathway is affected, leading to feedback regulation of the upstream pathways including photosynthesis, glycolysis, and the pentose phosphate pathway. This process decreases the flow of carbon sources into the shikimate pathway, resulting in decreased levels of secondary metabolites such as flavonoids, which ultimately affects plant growth and development (Fig. 7B). This model shows the regulation mechanism of BY1 on the biomass and grain yield of sorghum, highlighting its potential use for the future molecular design and breeding of sorghum, and it provides important insights into the relationship between plant development and metabolism.
Fig. 7.
A working model of the role of BY1 in regulating sorghum growth and development. (A) In the wild-type (WT), a portion of carbon dioxide fixed during photosynthesis is transformed into phosphoenolpyruvate (PEP) and erythritol-4-phosphate (E4-P) through glycolysis and the pentose phosphate pathway, respectively. PEP and E4-P enter the shikimate pathway under the control of BY1, pass through aromatic amino acid (AAA) biosynthesis and the flavonoid metabolism pathway, and form flavonoids, which regulate plant growth and development. (B) In the BY1 loss-of-function mutant (by1), the shikimate pathway is weakened, and feedback regulation leads to a decrease in the flow of carbon source the shikimate pathway, which leads to abnormal contents of secondary metabolites, thereby adversely affecting plant growth and development. The widths of the black arrows represent the amount of the carbon flow in the pathway. Scale bars in the images are 10 cm.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Morphology of panicle neck internodes, grains, and roots of the wild-type and by1 mutant.
Fig. S2. Morphology leaf veins of the wild-type and by1 mutant.
Fig. S3. Phenotypes of young panicles in the wild-type and by1 mutant before flowering.
Fig. S4. Phenotypes of fertile florets in the wild-type and by1 mutant.
Fig. S5. Sequencing and alignment of the mutation site among recessive individual plants, the by1 mutant, and the wild-type.
Fig. S6. Phylogenetic analysis of BY1 and its homologs.
Fig. S7. Phenotypic comparison between wild-type rice ZH11 and plants genome-edited plants expressing by1.
Fig. S8. Sequencing of BY1.
Fig. S9. Transcriptome analysis of BY1.
Fig. S10. Details of DEGs related to photosynthesis, phenylpropane metabolism, and the cell cycle.
Fig. S11. Details of DEGs related to nucleic acids, and mitosis and meiosis, together with RT-qPCR verification.
Fig. S12. RPKM values for genes related to glycolysis, and the pentose phosphate, shikimate, and phenylpropanoid pathways.
Fig. S13. Metabolic profiling of BY1.
Fig. S14. Metabolic map of pathways related to BY1.
Fig. S15. Comparison of the tertiary structures of BY1 in the wild-type and the by1 mutant.
Table S1. Comparison of agronomic traits between the wild-type and the by1 mutant.
Table S2. Characterization of metabolites present at different levels in the wild-type and the by1 mutant.
Table S3. Primers used in this study.
Data S1. List of sorghum varieties used to verify the mutation site in by1.
Data S2. Details of the 1785 DEGs identified between wild-type and the by1 mutant.
Acknowledgements
This research was supported by The National Key R&D Program of China (2018YFD1000706/2018YFD1000700), the China Postdoctoral Science Foundation (2018M642192) and The National Nature Science Foundation of China (3171444).
Author contributions
HC and JY designed and supervised this project; JC, MZ, RL, MZ, YL, YL, and XX performed the experiments; JC and HC analysed the data and wrote the manuscript.
Conflict of interest statement
The authors declare that they have no conflict of interest.
References
- Aharoni A, Galili G. 2011. Metabolic engineering of the plant primary–secondary metabolism interface. Current Opinion in Biotechnology 22, 239–244. [DOI] [PubMed] [Google Scholar]
- Anami SE, Zhang LM, Xia Y, Zhang YM, Liu ZQ, Jing HC. 2015. Sweet sorghum ideotypes: genetic improvement of the biofuel syndrome. Food and Energy Security 4, 159–177. [Google Scholar]
- Arcuri HA, Canduri F, Pereira JH, da Silveira NJ, Camera Júnior JC, de Oliveira JS, Basso LA, Palma MS, Santos DS, de Azevedo Júnior WF. 2004. Molecular models for shikimate pathway enzymes of Xylella fastidiosa. Biochemical and Biophysical Research Communications 320, 979–991. [DOI] [PubMed] [Google Scholar]
- Bentley R, Haslam E. 1990. The shikimate pathway — a metabolic tree with many branches. Critical Reviews in Biochemistry 25, 307–384. [DOI] [PubMed] [Google Scholar]
- Bickel H, Schultz G. 1979. Shikimate pathway regulation in suspensions of intact spinach chloroplasts. Phytochemistry 18, 498–499. [Google Scholar]
- Brunetti C, Di Ferdinando M, Fini A, Pollastri S, Tattini M. 2013. Flavonoids as antioxidants and developmental regulators: relative significance in plants and humans. International Journal of Molecular Sciences 14, 3540–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SA, Richards TA, Mui EJ, Samuel BU, Coggins JR, McLeod R, Roberts CW. 2004. A complete shikimate pathway in Toxoplasma gondii: an ancient eukaryotic innovation. International Journal for Parasitology 34, 5–13. [DOI] [PubMed] [Google Scholar]
- Chávez-Béjar MI, Lara AR, López H, Hernández-Chávez G, Martinez A, Ramírez OT, Bolívar F, Gosset G. 2008. Metabolic engineering of Escherichia coli for L-tyrosine production by expression of genes coding for the chorismate mutase domain of the native chorismate mutase-prephenate dehydratase and a cyclohexadienyl dehydrogenase from Zymomonas mobilis. Applied and Environmental Microbiology 74, 3284–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, Yu S, Xiong L, Luo J. 2013. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Molecular Plant 6, 1769–1780. [DOI] [PubMed] [Google Scholar]
- Cifuentes R, Bressani R, Rolz C. 2014. The potential of sweet sorghum as a source of ethanol and protein. Energy for Sustainable Development 21, 13–19. [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Research 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe T, Servos S, Hawkins A, Dimitriadis G, Young D, Dougan G, Charles I. 1991. The Mycobacterium tuberculosis shikimate pathway genes: evolutionary relationship between biosynthetic and catabolic 3-dehydroquinases. Molecular & General Genetics 228, 385–392. [DOI] [PubMed] [Google Scholar]
- Gilbert N. 2009. Averting a climate-led food crisis in Africa. Nature doi: 10.1038/news.2009.585. [DOI] [Google Scholar]
- Hay ME. 1996. Marine chemical ecology: what’s known and what’s next? Journal of Experimental Marine Biology and Ecology 200, 103–134. [Google Scholar]
- Herrmann KM. 1995. The shikimate pathway pathway: early steps in the biosynthesis of aromatic compounds. The Plant Cell 7, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann KM, Weaver LM. 1999. The shikimate pathway. Annual Review of Plant Physiology and Plant Molecular Biology 50, 473–503. [DOI] [PubMed] [Google Scholar]
- Jensen RA. 2006. The shikimate/arogenate pathway: link between carbohydrate metabolism and secondary metabolism. Physiologia Plantarum 66, 164–168. [Google Scholar]
- Keith B, Dong XN, Ausubel FM, Fink GR. 1991. Differential induction of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase genes in Arabidopsis thaliana by wounding and pathogenic attack. Proceedings of the National Academy of Sciences, USA 88, 8821–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn BM, Geisler M, Bigler L, Ringli C. 2011. Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiology 156, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Walbot V. 2007. Translational genomics for bioenergy production from fuelstock grasses: maize as the model species. The Plant Cell 19, 2091–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ML, Yuyama N, Luo L, Hirata M, Cai H. 2009. In silico mapping of 1758 new SSR markers developed from public genomic sequences for sorghum. Molecular Breeding 24, 41–47. [Google Scholar]
- Light SH, Anderson WF. 2013. The diversity of allosteric controls at the gateway to aromatic amino acid biosynthesis. Protein Science 22, 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu H, Gong Y, et al. . 2017. An atypical thioredoxin imparts early resistance to sugarcane mosaic virus in maize. Molecular Plant 10, 483–497. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma N, Wei L, Fan Y, Hua Q. 2012. Heterologous expression and characterization of soluble recombinant 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Actinosynnema pretiosum ssp. auranticum ATCC31565 through co-expression with Chaperones in Escherichia coli. Protein Expression and Purification 82, 263–269. [DOI] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. 2012. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annual Review of Plant Biology 63, 73–105. [DOI] [PubMed] [Google Scholar]
- Maeda H, Yoo H, Dudareva N. 2011. Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nature Chemical Biology 7, 19–21. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proceedings of the National Academy of Sciences, USA 88, 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazmi AR, Schofield LR, Dobson RC, Jameson GB, Parker EJ. 2014. Destabilization of the homotetrameric assembly of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from the hyperthermophile Pyrococcus furiosus enhances enzymatic activity. Journal of Molecular Biology 426, 656–673. [DOI] [PubMed] [Google Scholar]
- Pagor R, Sikdar MSL, Moon ES, Kim JS. 2015. Characterization of a gene encoding 3-deoxy-D-arabino-heptulosonate-phosphate synthase from rice. Journal of Biological Sciences 4, 461–466. [Google Scholar]
- Palakolanu SR, Dumbala SR, Kaliamoorthy S, Pooja BM, Vincent V, Sharma KK. 2016. Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Frontiers in Plant Science 7, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, et al. . 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556. [DOI] [PubMed] [Google Scholar]
- Rippert P, Scimemi C, Dubald M, Matringe M. 2004. Engineering plant shikimate pathway for production of tocotrienol and improving herbicide resistance. Plant Physiology 134, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts F, Roberts CW, Johnson JJ, et al. . 1998. Evidence for the shikimate pathway in apicomplexan parasites. Nature 393, 801–805. [DOI] [PubMed] [Google Scholar]
- Takaki M, Tan L, Murakami T, Tang YQ, Sun ZY, Morimura S, Kida K, Kida K. 2015. Production of biofuels from sweet sorghum juice via ethanol–methane two-stage fermentation. Industrial Crops and Products 63, 329–336. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. 2005. Flavonoids as developmental regulators. Current Opinion in Plant Biology 8, 317–323. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. . 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. [DOI] [PubMed] [Google Scholar]
- Tzin V, Galili G. 2010. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Molecular Plant 3, 956–972. [DOI] [PubMed] [Google Scholar]
- Tzin V, Malitsky S, Aharoni A, Galili G. 2009. Expression of a bacterial bi-functional chorismate mutase/prephenate dehydratase modulates primary and secondary metabolism associated with aromatic amino acids in Arabidopsis. The Plant Journal 60, 156–167. [DOI] [PubMed] [Google Scholar]
- Tzin V, Malitsky S, Ben Zvi MM, Bedair M, Sumner L, Aharoni A, Galili G. 2012. Expression of a bacterial feedback-insensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase of the shikimate pathway in Arabidopsis elucidates potential metabolic bottlenecks between primary and secondary metabolism. New Phytologist 194, 430–439. [DOI] [PubMed] [Google Scholar]
- Wang Y, Herrmann KM, Weller SC, Goldsbrough PB. 1991. Cloning and nucleotide sequence of a complementary DNA encoding 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from tobacco. Plant Physiology 97, 847–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Xu Z. 2019. Sustainable agriculture: from sweet sorghum planting and ensiling to ruminant feeding. Molecular Plant 12, 603–606. [DOI] [PubMed] [Google Scholar]
- Yamada T, Matsuda F, Kasai K, Fukuoka S, Kitamura K, Tozawa Y, Miyagawa H, Wakasa K. 2008. Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. The Plant Cell 20, 1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemaru J, Ando T, Mizubayashi T, Kasuga S, Matsumoto T, Yano M. 2009. Development of genome-wide simple sequence repeat markers using whole-genome shotgun sequences of sorghum (Sorghum bicolor (L.) Moench). DNA Research 16, 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalza A, Orcaray L, Fernández-Escalada M, Zulet-González A, Royuela M. 2017. The pattern of shikimate pathway and phenylpropanoids after inhibition by glyphosate or quinate feeding in pea roots. Pesticide Biochemistry and Physiology 141, 96–102. [DOI] [PubMed] [Google Scholar]
- Zhang WN, He JJ, Pan QH, Han FL, Duan CQ. 2011. Separation and character analysis of anthocyanins from mulberry (Morus alba L.) pomace. Czech Journal of Food Sciences 29, 268–276. [Google Scholar]
- Zhao J, Herrmann KM. 1992. Cloning and sequencing of a second cDNA encoding 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Solanum tuberosum L. Plant Physiology 100, 1075–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Wang S, Huang Z, et al. . 2018. Rewiring of the fruit metabolome in tomato breeding. Cell 172, 249–261.e12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.