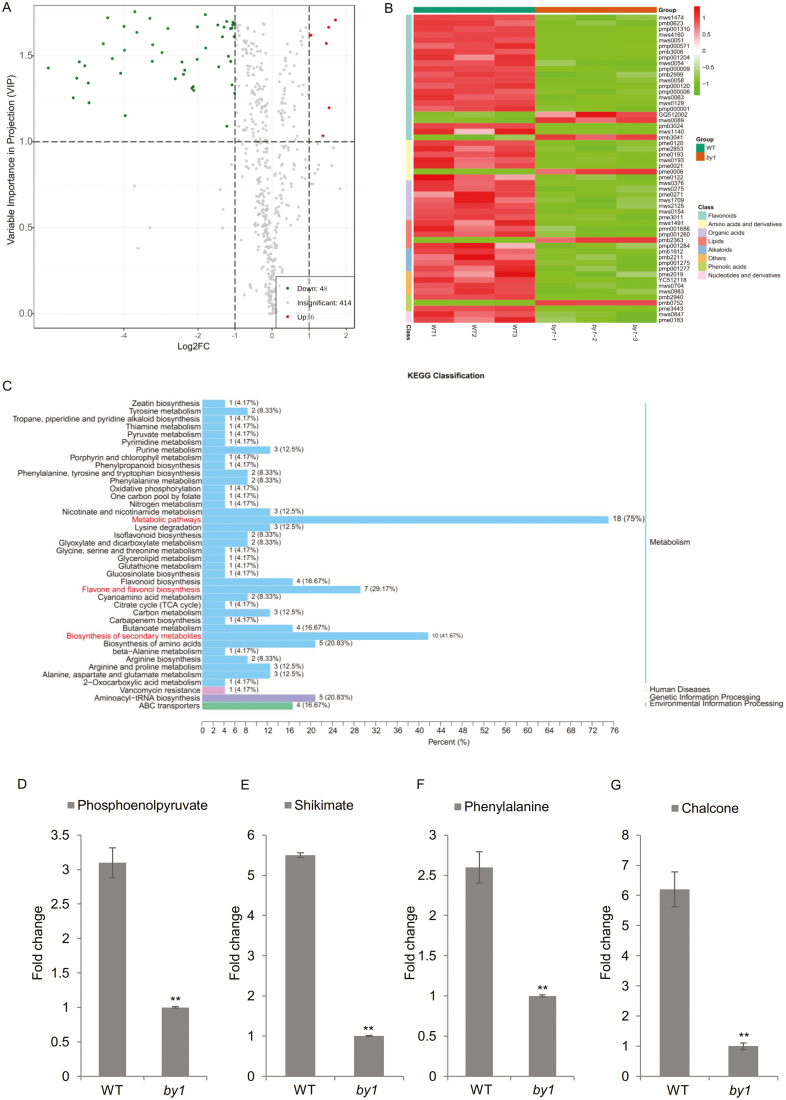
Fig. 6.
Metabolic profiling analysis of sorghum wild-type BY1 and the by1 mutant. (A) Volcano plot of the different metabolites. For each metabolite, the variable importance in projection (VIP) value is plotted against the logarithm of the quantitative difference multiple of the metabolite in two samples: the larger the absolute value, the greater the difference multiple of the content of the metabolite. The larger the VIP value, the more significant the difference in the metabolite levels. Metabolites down-regulated in by1 relative to the wild-type (WT) are shown in green, metabolites up-regulated are shown in red, and those with no significant difference are shown in gray. (B) Clustering heatmap of metabolites with significantly different contents (MSDCs) in by1 compared with the WT. The metabolite classes are grouped according to the color scale and their contents are indicated by the red–green scale. WT lines are to the left and by1 lines are to the right. (C) KEGG classification of MSDCs. The number of the MSDCs annotated to each pathway is shown together with its percentage of the total number of MSDCs. (D–G) Fold-changes of (D) phosphoenolpyruvate, (E) shikimate, (F) phenylalanine, and (G) chalcone levels between the WT and by1. Data are means (±SD), n=3. Significant differences were determined using two-tailed Student’s t-test: **P<0.01.