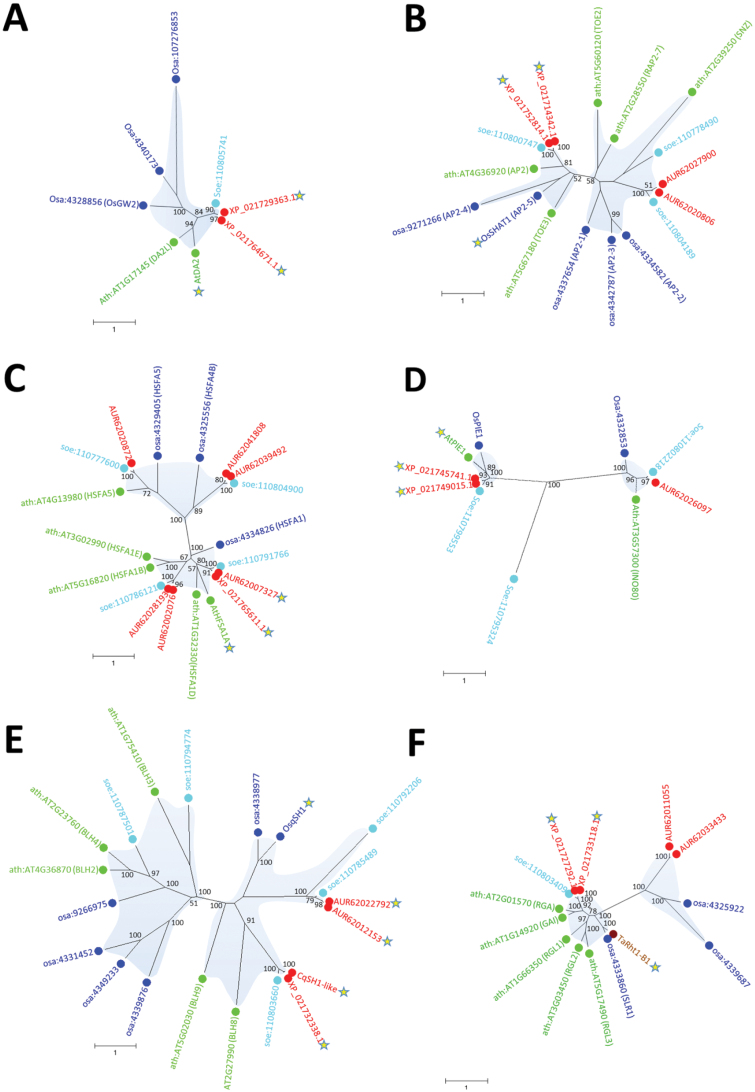
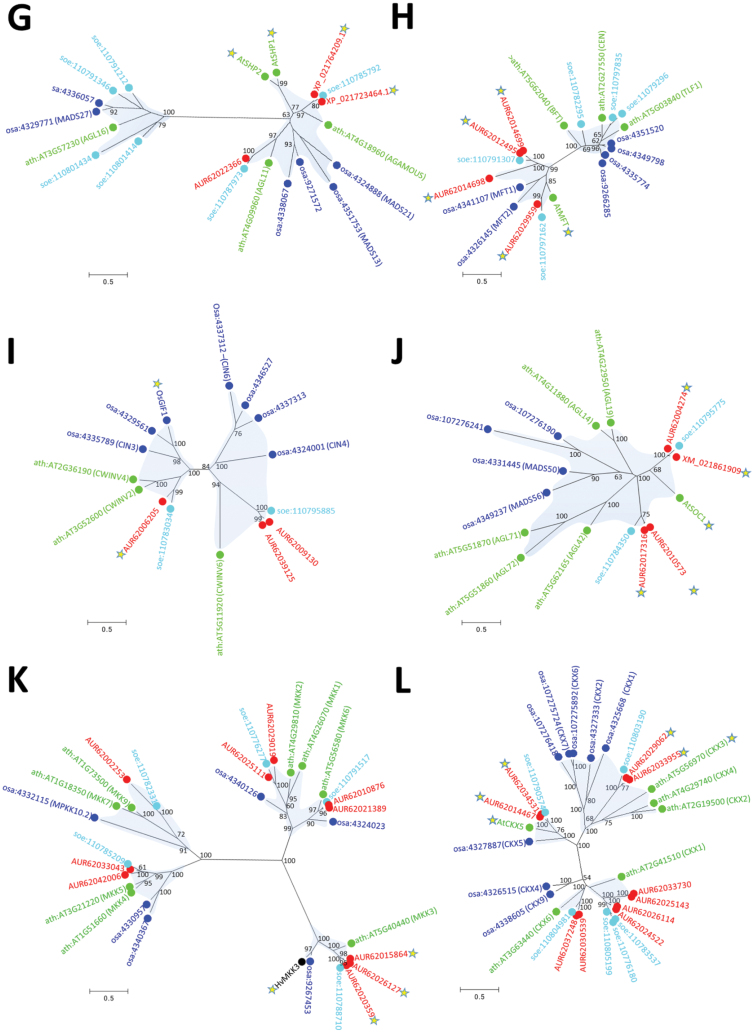
Fig. 2.
Phylogenetic tree of gene families in which members from rice (Oryza sativa), wheat (Triticum aestivum), and Arabidopsis thaliana control traits are suggested to be important for domestication of quinoa. Homologous genes in spinach (Spinachia oleracea), which is closely related to quinoa, are also shown. Species origins are highlighted by coloured text and circles: red, quinoa; blue, rice; green, Arabidopsis; turquoise, spinach; brown, wheat; black, barley (Hordeum vulgare). Domestication genes and their closest homologues in quinoa are marked by yellow stars. (A) OsGW2 controls seed size in rice. (B) OsSHAT1 controls seed shattering in rice. (C) AtHFSA1A controls heat stress in Arabidopsis. (D) OsPIE1 controls flowering time in rice. (E) OsqSH1 controls seed shattering in rice. (F) TaRht1-B1 controls plant height in wheat. (G) AtSHP1 controls seed dispersal in Arabidopsis. (H) AtMFT controls early sprouting in Arabidopsis. (I) OsGIF1 is involved in seed size in rice. (J) AtSOC1 controls flowering time in Arabidopsis. (K) HvMKK3 controls seed dormancy in barley. (L) Loss-of-function double mutation of AtCKX5 and AtCKX3 in Arabidopsis mimics the rice gn1a mutation related to increased grain numbers. (M) AtTFL1 is a time-of-flowering regulator in Arabidopsis and other species. For references, see main text. Accession numbers not given in the figure are as follows: AtDA2, Q93YV5; OsGIF1, Q6AVI1; OsGW2, B9F4Q9; AtMFT, Q6XFK7; AtHSFA1A, P41151; AtPIE1, Q7X9V2; OsqSH1, Q941S9; TaRHT1, Q9ST59; OsSHAT1, A0A0N7KJT8; AtSHP1, P29381; AtSHP2, P29385; AtSOC1, O64645; HvMKK3, A0A140JZ28; AtCKX5, Q67YU0; and AtCKX3, A0A1P8BER3. The basic local alignment search tool (BLAST) was used to search for genes in genomes annotated in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/tools/blast/), the KAUST Chenopodium database (https://www.cbrc.kaust.edu.sa/chenopodiumdb/), and the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/?term=quinoa). CqSH1-like (previously AUR62029222) was not correctly annotated and was corrected based on homology to the coding sequences of OsQSH1 and soe:110803660 guided by intron–exon splice sites in the quinoa genome sequence. The sequences were aligned using the multiple sequence comparison by the log-expectation (MUSCLE; Edgar, 2004) tool and subjected to maximum likelihood analysis by RAxML v. 8.2.12 (Stamatakis, 2014) assuming a Le and Gascuel (LG)+PROTGAMMA model (Le and Gascuel, 2008) and using the Extreme Science and Engineering Discovery Environment (XSEDE) at the CIPRES Science Gateway v. 3.3 (Miller et al., 2010). Bootstrap values from 1000 replicates are indicated on each node. Values <50 are not marked. Scale bars have numbers of amino acid substitutions per site indicated below.