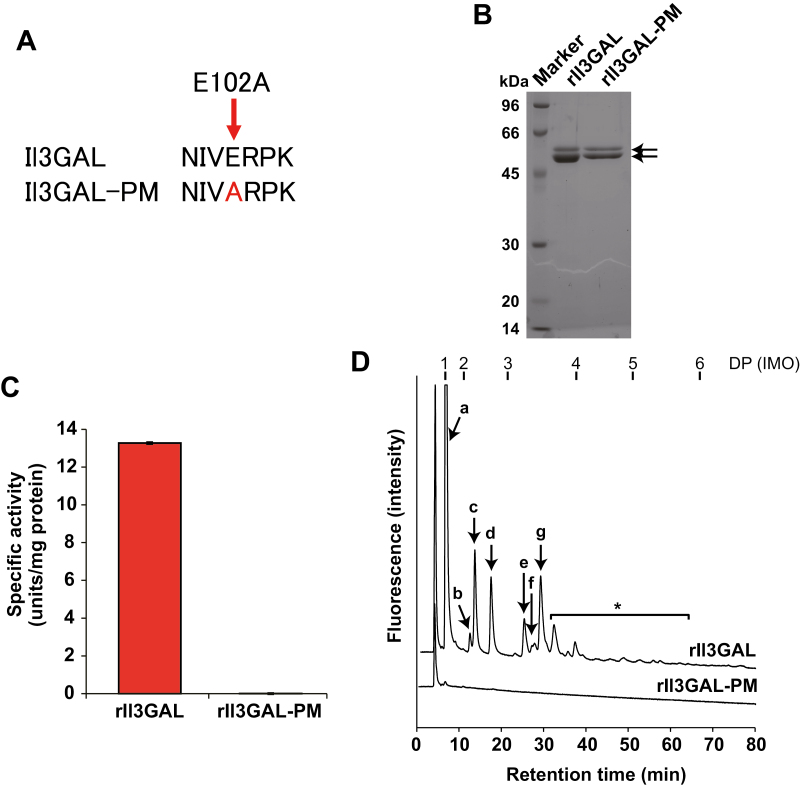
Fig. 2.
Enzymatic activity of rIl3GAL and rIl3GAL-PM. (A) Point mutation in the catalytic site of Il3GAL. The Glu residue was replaced with Ala to generate rIl3GAL-PM. (B) The purity of recombinant proteins. The rIl3GAL and rIl3GAL-PM proteins were examined by SDS–PAGE. Proteins in the gel were stained with Coomassie Brilliant Blue R-250. Arrows indicate rIl3GAL and rIl3GAL-PM. (C) Galactanase activity. The activity of recombinant enzymes was measured using β-1,3-galactan as substrate. Data are mean values ±SD (n=3 technical replicates). (D) Released oligosaccharides from radish leaf AGPs by the action of rIl3GAL or rIl3GAL-PM. Enzymatic hydrolysis products were derivatized with ABEE and detected by HPLC. Arrows indicate oligosaccharides as follows: a, Gal; b, MeGlcAGal; c, Gal2; d, l-Ara-β-1,6-Gal2; e, MeGlcAGal2; f, Gal3; g, AraGal3. Asterisks indicate oligosaccharides released from type II AGs but not assigned. The elution positions of glucose and IMOs with degree of polymerization (DP) 2–6 are indicated on the top.