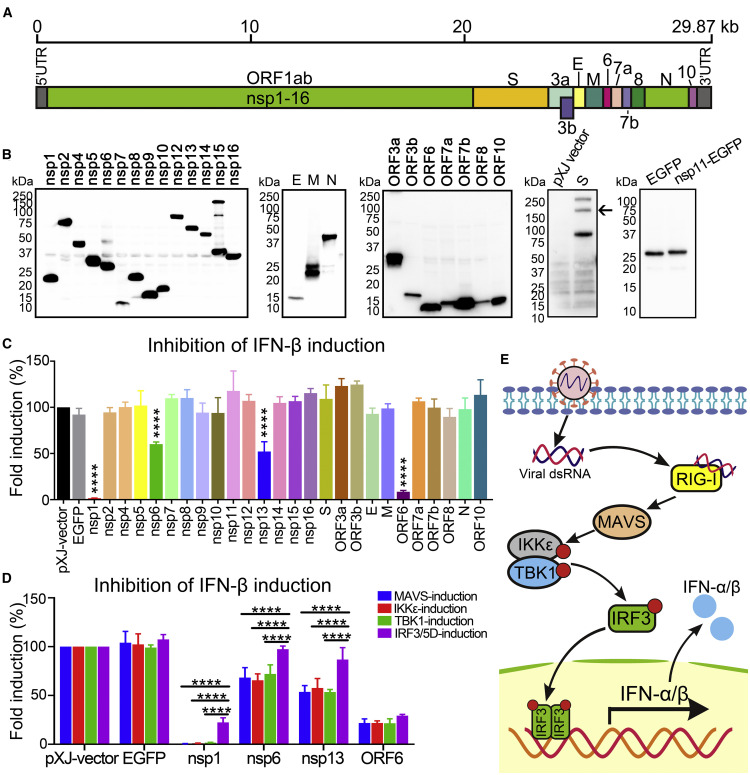
Figure 1.
SARS-CoV-2 Proteins Inhibit IFN-I Production
(A) Genome structure of SARS-CoV-2.
(B) Expression of SARS-CoV-2 proteins. C-Terminally FLAG-tagged viral proteins were expressed in HEK293T cells and analyzed by western blotting using anti-FLAG antibody. S protein was probed by anti-S antibody and is indicated by an arrow; an empty pXJ-plasmid-transfected cell lysate was included as a negative control. EGFP was fused to the C terminus of nsp11 and probed by anti-GFP antibody. All viral proteins were cloned from SARS-CoV-2 strain 2019-nCoV/USA_WA1/2020.
(C) IFN-β promoter luciferase assay. HEK293T cells were co-transfected with Firefly luciferase reporter plasmid pIFN-β-luc, Renilla luciferase control plasmid phRluc-TK, viral protein expressing plasmid, and stimulator plasmid RIG-I (2CARD). Empty plasmid and EGFP-encoding plasmid were used as controls. Cells were assayed for luciferase activity at 24 hpt. The data were analyzed by normalizing the Firefly luciferase activity to the Renilla luciferase (Rluc) activity and then normalized by non-stimulated samples to obtain fold induction. Empty vector control was set to 100%. Statistics were determined by comparing with EGFP control and one-way ANOVA with Dunnett’s correction, ∗∗∗∗p < 0.0001.
(D) MAVS-, IKKε-, TBK1-, or IRF3/5D-activated IFN-β promoter luciferase assay. The experiments were performed as in (C) except that the assay was activated by MAVS, IKKε, TBK1, or IRF3/5D. Data were from three independent experiments in triplicate (mean ± SD). Statistics were determined by comparing each respective IRF3/5D-induction group using two-way ANOVA with Dunnett’s correction, ∗∗∗∗p < 0.0001.
(E) Scheme of RIG-I-mediated IFN-I production pathway.