Abstract
The outbreaks of the infectious disease COVID-19 caused by SARS-CoV-2 seriously threatened the life of humans. A rapid, reliable and specific detection method was urgently needed. Herein, we reported a contamination-free visual detection method for SARS-CoV-2 with LAMP and CRISPR/Cas12a technology. CRISPR/Cas12a reagents were pre-added on the inner wall of the tube lid. After LAMP reaction, CRISPR/Cas12a reagents were flowed into the tube and mixed with amplicon solution by hand shaking, which can effectively avoid possible amplicon formed aerosol contamination caused by re-opening the lid after amplification. CRISPR/Cas12a can highly specific recognize target sequence and discriminately cleave single strand DNA probes (5′-6FAM 3′-BHQ1). With smart phone and portable 3D printing instrument, the produced fluorescence can be seen by naked eyes without any dedicated instruments, which is promising in the point-of-care detection. The whole amplification and detection process could be completed within 40 min with high sensitivity of 20 copies RNA of SARS-CoV-2. This reaction had high specificity and could avoid cross-reactivity with other common viruses such as influenza virus. For 7 positive and 3 negative respiratory swab samples provided by Zhejiang Provincial Center for Disease Control and Prevention, our detection results had 100% positive agreement and 100% negative agreement, which demonstrated the accuracy and application prospect of this method.
Keywords: SARS-CoV-2, RT-LAMP, CRISPR/Cas12a, Visual detection
Abbreviations: qRT-PCR, quantitative Reverse Transcription-Polymerase Chain Reaction; RT-LAMP, Reverse Transcription-Loop-mediated isothermal amplification; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeat; dNTP, deoxyribonucleoside triphosphate; LOD, Limit of detection
Highlights
-
•
A contamination-free visual detection method of SARS-CoV-2 with CRISPR/Cas12a is developed.
-
•
The detection process can be completed within 40 min with high sensitivity of as low as 20 copies RNA.
-
•
The detection results can be observed by our portable 3D printing instrument and smart phone.
-
•
For real respiratory swab samples detection, our detection results have 100% positive agreement and 100% negative agreement.
1. Introduction
A severe respiratory disease caused by SARS-CoV-2 was found in December 2019 (Wu et al., 2020a). The familial clustered cases suggested that the virus could spread from person to person (Chan et al., 2020). It was found that the infectivity of SARS-CoV-2 peaked during the incubation period, which may cause many clustered cases (He et al., 2020). Diagnosis in time is effective for tracking and controlling the epidemic. The commonly used testing methods include immunoassays and nucleic acid detection. However, for immunoassays, it was difficult to detect blood samples in the incubation and early infection period. Cross-reaction with other similar coronaviruses may also affect the specificity of immunoassays (Lee et al., 2020). Therefore, immunoassays could not be used as the only basis but a supplement for the diagnosis and exclusion of COVID-19. qRT-PCR was the gold standard for SARS-CoV-2 detection extracted from throat or nasal swabs (Wu et al., 2020c). However, qRT-PCR requires repeated heat and cold process with bulky and expensive equipment. The complex operation steps of qRT-PCR testing must be done by special labs or hospitals, which is difficult to detect a large number of samples in such a short time. For some low-resource areas, diagnosis of COVID-19 by qRT-PCR is more difficult.
For RT-LAMP reaction, thermo-cycling process is not required and the reaction time is also shorter than qRT-PCR (Estrela et al., 2019). It is more suitable for point-of care detection. Researchers are constantly exploring new detection technologies. CRISPR technology is a highly specific tool for nucleic acid detection. Zhang et al. developed Cas13-based SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) technology to detect Zika virus and dengue virus with high sensitivity and specificity (Myhrvold et al., 2018). Doudna et al. found the Cas12a could cleave single strand DNA indiscriminately when it bound to the target sequence under the guidance of gRNA. Based on it, they developed DETECTR (DNA endonuclease-targeted CRISPR trans reporter) method to detect the virus (Chen et al., 2018). With the spread of the global epidemic, more rapid and convenient detection methods are being explored. Based on CRISPR system and lateral flow assays, Broughton et al. reported a rapid method that could detect SARS-CoV-2 within 40 min. Extracted RNA from the respiratory swab sample were amplified by RT-LAMP. Then the tube was open and CRISPR/Cas12 reagents were added into the amplicon solution. Finally the detection results were read out by inserting lateral flow strips into the tube (Broughton et al., 2020). This detection method stepped further towards point-of-care testing. However, after LAMP amplification the concentration of the target nucleic acids in the tube was high. The amplicons may form aerosol contamination when re-opening the lid, which was a serious problem for nucleic acid detection. PCR has been widely used in molecular diagnosis since it was first published. The limited studies showed that false-positive rates in the laboratory was high because of contamination (Borst et al., 2004). Due to high sensitivity of nucleic acid amplification such as PCR and LAMP, DNA fragments from the previous experiments may be re-amplified, causing the false positive results of the next detection (Mukama et al., 2020). To prevent amplicons formed aerosol contamination, our research group had done some work based on CRISPR/Cas technology (Qian et al., 2019; Wu et al., 2020b).
Here, we reported a contamination-free visual method for SARS-CoV-2 detection with CRISPR/Cas12a. Having extracted RNA from respiratory throat swab sample, RT-LAMP reagents were added in the tube. Mineral oil was added to cover the RT-LAMP reagents, which could prevent volatilization of reaction solution and diffusion of amplicons. CRISPR/Cas12a reaction liquid was pre-added on the inner wall of the tube lid. Even if the lid was closed, the CRISPR/Cas12a reagents would not drip because of the surface tension. The tube was put at the isothermal condition to complete RT-LAMP reaction. Then shaking the tube to make the pre-added CRISPR/Cas12a reagents flow into it rather than re-open the tube to add reagents, which can further avoid possible amplicons formed aerosol contamination caused by opening the lid. Cas12a protein recognize the target nucleic acids and discriminately cleave the single strand DNA probes (5′-6FAM 3′-BHQ1). The tube was finally put in the 3D printing instrument and the fluorescent signal could be seen by the naked eyes with the smart phone. We have demonstrated this method had high specificity and could prevent cross-reaction with other common viruses such as influenza virus. By covering a layer of mineral oil and pre-adding CRISPR reagents, the possible amplicon formed aerosol contamination could be effectively prevented. Combined with the portable 3D printing instrument, the detection results could be judged by producing fluorescence or not. No bulky and expensive instruments were needed in the whole testing process. We anticipate this detection method is promising in the point-of-care detection for SARS-CoV-2.
2. Materials and methods
2.1. Materials
Synthesized RNA templates were purchased from Genewell Gene Technology Co., Ltd, (Shenzhen, China). RNA templates were diluted by a 10-fold gradient and the concentrations were 100, 101, 102, 103, 104 copies/μL. The real SARS-CoV-2 samples were extracted and provided by Zhejiang Provincial Center for Disease Control and Prevention. Primers of qRT-PCR, RT-LAMP, qRT-PCR probes, CRISPR probes and CRISPR gRNA were synthesized by Sangon Biotech (Shanghai, China).
2.2. qRT-PCR assay
The qRT-PCR assay was performed in a 25 μL system. qRT-PCR primers of ORF gene of SARS-CoV-2 were designed according to the recommendations of Chinese Center for Disease Control and Prevention. qRT-PCR assay was conducted by One Step PrimeScript™ RT-PCR Kit (TaKara Biomedical Technology Co., Ltd., Beijing, China). The reaction system contains 1 × One Step RT-PCR Buffer Ⅲ, 0.08 U/μL TaKaRa Ex Taq HS, 0.2 μM Forward Primer, 0.2 μM Reverse Primer, 0.1 μM probe, 0.4 μL PrimeScript RT Enzyme Mix Ⅱ, 2 μL RNA templates of different concentrations. The temperature procedure of qRT-PCR was 42 °C for 5 min, 95 °C for 10 s, 40 cycles at 95 °C for 5 s and 60 °C for 30 s. The amplification process was conducted in StepOne™ Plus Real-Time PCR System (Applied Biosystems, Inc., Carlsbad, CA, USA).
2.3. Real-time RT-LAMP assay
The real-time RT-LAMP assay was performed in a 25 μL system. The reaction mixture contained 1 × Isothermal Amplification Buffer, 0.32 U/μL Bst 2.0 DNA polymerase (New England Biolabs Inc., Ipswich, MA, USA), 1.4 mM dNTPs (New England Biolabs Inc., Ipswich, MA, USA), 8 mM MgSO4 (New England Biolabs Inc., Ipswich, MA, USA), 1.6 μM FIP primers, 1.6 μM BIP primers, 0.2 μM F3 primers F3, 0.2 μM B3 primers, 0.4 μM LF primers, 0.4 μM LB primers, 0.3 U/μL WarmStart® RTx Reverse transcriptase (New England Biolabs Inc., Ipswich, MA, USA), 1 × SYBR Green I (Thermo Fisher Scientific, Inc., Waltham, MA, USA), 2 μL RNA templates of different concentrations. Real-time RT-LAMP assays of ORF gene, N gene and E gene were performed using the same reaction system except for different designed primers. 35 μL of mineral oil was covered on the real-time RT-LAMP reaction solution. Real-time RT-LAMP assay was performed at 65 °C for 40 min in StepOne™ Plus Real-Time PCR System (Applied Biosystems, Inc., Carlsbad, CA, USA).
2.4. Contamination-free visual detection assay with CRISPR/Cas12a
35 μL of mineral oil was covered on the RT-LAMP reaction solution (The reaction system was the same as real time RT-PCR except that the fluorescent dyes were replaced with water). After RT-LAMP reaction, 20 μL of CRISPR reaction solution (pre-added inside the lid) was mixed with the RT-LAMP amplification solution by hand shaking. 20 μL CRIPSR reaction solution contained 1 × NEBuffer 2.1 Reaction Buffer, 0.2 μM EnGen® Lba Cas12a (Cpf1) (New England Biolabs Inc., Ipswich, MA, USA), 0.6 μM gRNA, 2.5 μM CRISPR probes, 1 U/μL RNA inhibitor (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The tube was put in ProFlex PCR System (Applied Biosystems, Inc., Carlsbad, CA, USA) at 37 °C for 5 min. After the reaction, 3D printing instrument and smart phone were used to observe the fluorescence.
2.5. The specificity assay and real SARS-CoV-2 sample detection
In order to verify the specificity of our method, we selected common viruses associated with respiratory infections to detect. They were Conoravirus 229E, Conoravirus NL63, Conoravirus OC43, Conoravirus HKU1, Influenza virus A (FluA), Influenza virus B (FluB), Human Adenovirus, Human respiratory syncytial virus A (RSV-A), Human RSV-B, Human parainfluenza virus1 (PIV1), Human PIV2, Human PIV3, Human PIV4, Human Metapneumovirus (hMPV), Human Bocavirus, Human enterovirus, Human rhinovirus, Polyomavirus (PyV) (Zhejiang Provincial Center for Disease Control and Prevention). We used our contamination-free visual detection method to detect real SARS-CoV-2 and other virus samples. These real samples were extracted by Zhejiang Provincial Center for Disease Control and Prevention. To protect the privacy of the patients, these real samples were re-numbered.
3. Results and discussion
3.1. The design of primers for RT-LAMP and gRNA for CRISPR reaction
The gene sequence of the SARS-CoV-2 was about 30 kb. Two thirds of the viral genome RNA sequence were open reading frames (ORF). It was reported that the structural proteins of SARS-CoV-2 included the envelope protein (encoded by E gene), the membrane protein (encoded by M gene), the nucleocapsid protein (encoded by N gene) and the spike protein (encoded by S gene) (Kim et al., 2020). Based on the sequencing and transcriptome analysis, three gene fragments, ORF gene, N gene and E gene were selected for detection (Fig. 1 ). Sequence information of primers, probes and CRISPR gRNA were summarized in Table 1 in the supplementary information.
Fig. 1.
In the whole gene sequence of the SARS-CoV-2, the ORF gene, E gene, N gene were chosen as the targets for detection.
Table 1.
Other common 17 viruses were tested for the specificity assay of the contamination-free visual detection method.
| The specificity assay of the visual detection of SARS-CoV-2 | ||||
|---|---|---|---|---|
| Standard RNA sample | Conoravirus 229E 2016HZ-781 | Conoravirus 229E 2016HZ-803 | Conoravirus 229E 2016ZS-842 | Conoravirus 229E 2019HZ-688 |
| + | - | - | - | - |
| Conoravirus NL63 2018HZ-755 | Conoravirus NL63 2018HZ-797 | Conoravirus OC43 2019HZ-653 | Conoravirus OC43 2019ZS-802 | Conoravirus HKU1 2018ZS-537 |
| - | - | - | - | - |
| Conoravirus HKU1 2019ZS-106 | FluA 2019HZ-757 | FluB 2019ZS-807 | Human Adenovirus 2019GC-900 | Human RSV-A 2019LS-355 |
| - | - | - | - | - |
| Human RSV-B 2019LS-203 | Human PIV2 2019ZS-818 | Human PIV3 2019GC-907 | Human PIV4 2018ZS-742 | Human Metapneumovirus 2019LS- 309 |
| - | - | - | - | - |
| Human Bocavirus 2019GC- 911 | Human Enterovirus 2019ZS -816 | Human Rhinovirus 2019ZS -812 | polyomavirus PyV 2019LS-301 | polyomavirus PyV 2016HZ-796 |
| - | - | - | - | - |
3.2. qRT-PCR assays for RNA samples
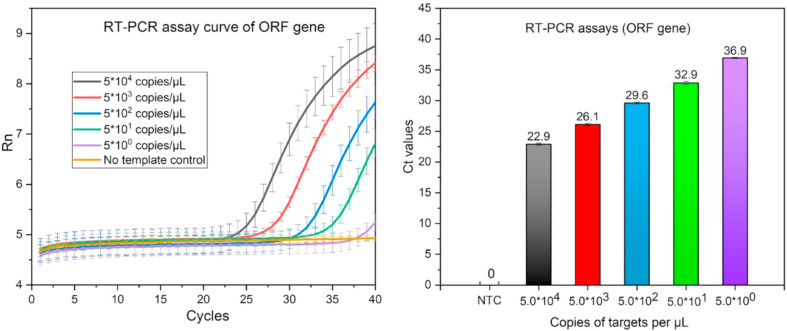
qRT-PCR assay of ORF gene of SARS-CoV-2 was conducted according to the recommendation from Chinese Center for Disease Control. According to the results in Fig. 2 , it could be seen that the samples with five concentration gradients could be detected with cycle threshold (Ct) values of 22.9, 26.1, 29.6, 32.9, 36.9 respectively. The qRT-PCR assay of ORF gene could detect as low as 2 copies RNA of SARS-CoV-2 per reaction. These experiments were repeated three times. qRT-PCR process must be conducted in bulky instrument with thermo-cycling and real time fluorescence signal acquisition ability, which limited its wider application.
Fig. 2.
The RT-PCR amplification curves and corresponding Ct values for ORF gene of different concentrations of RNA templates using methods recommended by Chinese Center for Disease Control and Prevention.
3.3. The real time RT-LAMP for E gene, N gene and ORF gene
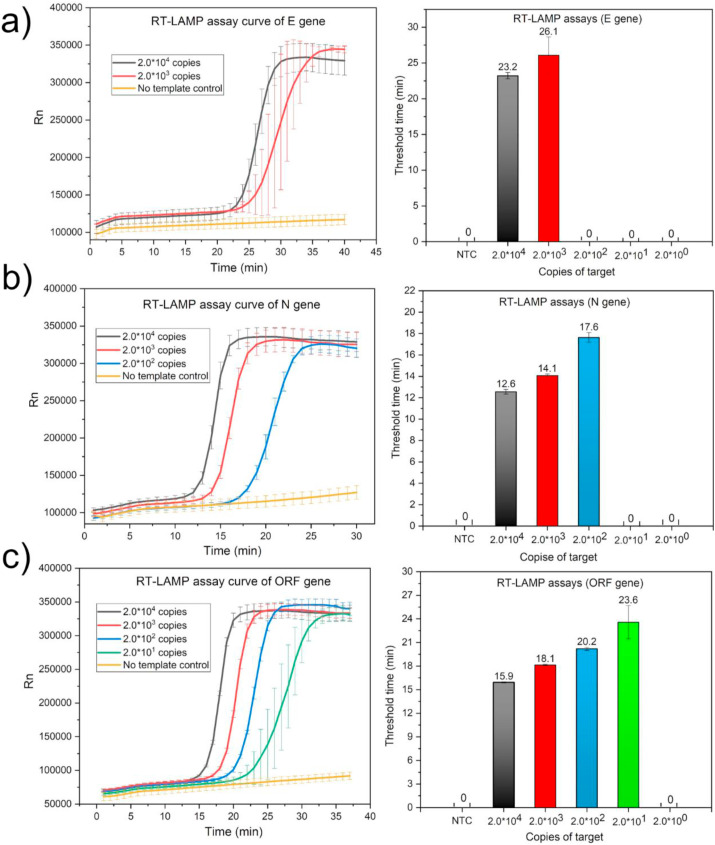
Real time RT-LAMP primers of E gene, N gene and ORF gene were designed. The target amplification sequences all contained protospacer adjacent motif (PAM) sequence for CRISPR/Cas detection. The detection results of real time RT-LAMP for E, N and ORF gene were showed in Fig. 3 . Among the real time RT-LAMP assays for these three genes, the LOD for ORF gene was the lowest with 20 copies per reaction. When the concentration of the target RNA was as low as 2 copies, real time RT-LAMP assays had no stable fluorescent signal. ORF gene was selected for the next CRISPR/Cas based visual detection. The real time RT-LAMP reaction could be completed within 40 min at 65 °C. Although the LOD of the real time RT-LAMP assays was an order of magnitude lower than that of qRT-PCR, the advantage of the RT-LAMP was that it did not require repeated heating and cooling process. RT-LAMP had lower requirements for the instruments so it was more suitable for the point-of-care detection.
Fig. 3.
The real time RT-LAMP amplification curves and corresponding threshold time of different concentrations of RNA templates with a) E gene as the target. b) N gene as the target. c) ORF gene as the target.
3.4. Contamination-free visual detection with CRISPR/Cas12a
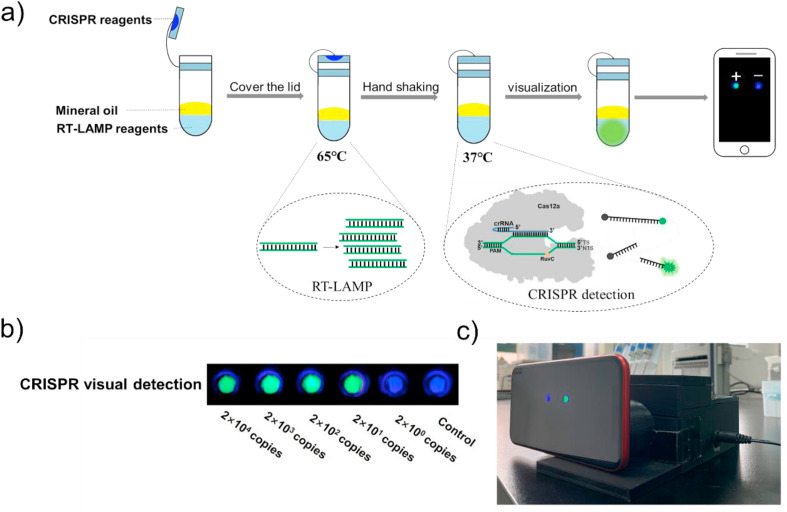
Compared with real time detection, endpoint detection was easier to realize in the point-of-care testing. However, after LAMP reaction, if the lid of tube was re-open for the next detection it may form amplicon aerosol contamination carrying target nucleic acids, which may cause false positive results in the next detection. Here, we introduced CRIPSR technology to realize the endpoint visual detection of SARS-CoV-2. As illustrated in Fig. 4 a, mineral oil was added to cover the extracted nucleic acids and RT-LAMP solution. CRISPR/Cas12a reagents were pre-added inside of the tube lid. After RT-LAMP reaction, CRISPR/Cas12a reagents were mixed with amplification reagents by hand shaking. The whole process did not need to open the lid twice, avoiding the amplicon aerosol contamination. As you could see the results in Fig. 4b, there were green fluorescence when the concentrations of RNA samples (ORF gene as the target fragments) were 2 × 104, 2 × 103, 2 × 102, 2 × 101 copies. For sample of 2 × 100 copies and no template control group, there was no green fluorescence signal. The detection results were visualized using smart phone and portable 3D printing fluorescence detection instrument designed by us (Fig. 4c). The LOD of ORF gene was consistent with the above real time RT-LAMP amplification results.
Fig. 4.
a) The diagram of overall experimental flow and principle of the contamination-free visual detection method. b) Visual detection of different concentrations of synthesized RNA templates of ORF gene. c) The photo of 3D printing instrument and smart phone used to observe fluorescent signal.
3.5. Verification the specificity of the contamination-free visual detection method
Highly specific detection method can avoid cross-reactions and improve the accuracy of detection. To demonstrate the specificity of the contamination-free visual detection method, other common virus samples were detected. As was shown in Table 1, only synthesized RNA template of ORF gene produced green fluorescence, which demonstrated that our method had high specificity.
3.6. The visual detection of real samples of SARS-CoV-2
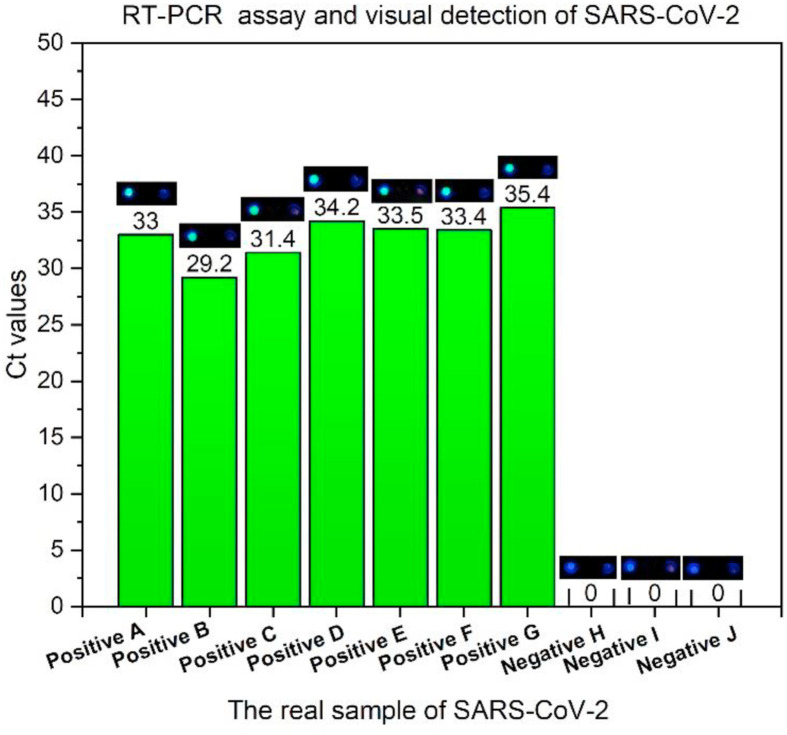
To demonstrate the feasibility of our method in practical application, 10 real samples provided by Zhejiang Provincial Center for Disease Control and Prevention were detected, of which 7 positive samples and 3 negative samples. ORF gene was the target fragments for detection. For these 7 positive samples, the Ct values of qRT-PCR were 33, 29.2, 31.4, 34.2, 33.5, 33.4, 35.4. These real samples were detected using our contamination-free visual detection method. As you could see in Fig. 5 , 7 positive samples all produced green fluorescence (left) and 3 negative samples did not produce green fluorescence. Relative to the qRT-PCR detection method recommended by CDC, our detection results had 100% positive agreement and 100% negative agreement. When the Ct value of positive G sample was 35.4, our contamination-free visual method could also detect it accurately, indicating that our method had high sensitivity even if the number of target RNA in the real sample was relatively small. Also, the entire process did not need to open the lid, which could effectively avoid false positive results caused by aerosol contamination.
Fig. 5.
The detection results of 7 positive and 3 negative real respiratory swab samples. For each picture, the left side was sample to be detected and the right side was no template control.
4. Conclusion
We reported a contamination-free visual method for SARS-CoV-2 detection with CRISPR/Cas12a. By covering mineral oil on LAMP solution and pre-adding CRISPR reagents in the tube lid, the entire process did not require opening the lid again, which could effectively avoid the false positive caused by amplicon formed aerosol contamination. RT-LAMP reaction did not involve repeated heating and cooling process. With CRISPR/Cas12a technology, the detection results could be seen directly on smart phones under the help of the 3D printing instrument, which greatly reduced the cost and increased the portability of testing. The LOD of ORF gene of SARS-CoV-2 could be as low as 20 copies RNA per reaction. The visual detection process could be completed within 40 min. Our detection method was proved to be highly specific by testing other 17 common viruses. 7 positive and 3 negative real respiratory swab samples were detected with 100% positive agreement and 100% negative agreement. This fast, low-cost, high-specificity and high-sensitivity method could avoid using thermo-cycling instruments, specialized detector and possible aerosol contamination. It was promising in the point-of-care detection of SARS-CoV-2.
CRediT authorship contribution statement
Yanju Chen: Formal analysis, Writing - original draft. Ya Shi: Formal analysis, Methodology. Yin Chen: Methodology, Investigation. Zhangnv Yang: Investigation. Hui Wu: Investigation, Validation. Zhihui Zhou: Investigation, Validation. Jue Li: Validation. Jianfeng Ping: Investigation. Luping He: Resources, Methodology. Hong Shen: Conceptualization, Supervision. Zhengxin Chen: Conceptualization, Supervision. Jian Wu: Conceptualization, Supervision, Methodology, Funding acquisition, Writing - review & editing. Yunsong Yu: Conceptualization, Supervision. Yanjun Zhang: Conceptualization, Supervision. Huan Chen: Conceptualization, Project administration, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31571918), the Science and Technology Department of Zhejiang Province, China (Nos. 2020C03123 and LGN20C050001), the Key Program of Logistics Research of China (BWS17J030), National Science and Technology Major Project for “Major New Drugs Innovation and Development” (2015zx09j15102-002).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112642.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Borst A., Box A.T.A., Fluit A.C. Eur. J. Clin. Microbiol. Infect. Dis. 2004;23:289–299. doi: 10.1007/s10096-004-1100-1. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Yuan S.F., Kok K.H., To K.K.W., Chu H., Yang J., Xing F.F., Liu J.L., Yip C.C.Y., Poon R.W.S., Tsoi H.W., Lo S.K.F., Chan K.H., Poon V.K.M., Chan W.M., Ip J.D., Cai J.P., Cheng V.C.C., Chen H.L., Hui C.K.M., Yuen K.Y. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E.B., Harrington L.B., Da Costa M., Tian X.R., Palefsky J.M., Doudna J.A. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrela P.F.N., Mendes G.D., de Oliveira K.G., Bailao A.M., Soares C.M.D., Assuncao N.A., Duarte G.R.M. J. Virol. Methods. 2019;271:113675. doi: 10.1016/j.jviromet.2019.113675. [DOI] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X.L., Wang J., Hao X.X., Lau Y.C., Wong J.S.Y., Guan Y.J., Tan X.H., Mo X.N., Chen Y.Q., Liao B.L., Chen W.L., Hu F.Y., Zhang Q., Zhong M.Q., Wu Y.R., Zhao L.Z., Zhang F.C., Cowling B.J., Li F., Leung G.M. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.Y.-P., Lin R.T.P., Renia L., Ng L.F.P. Front. Immunol. 2020;11:879. doi: 10.3389/fimmu.2020.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukama O., Nie C.R., Habimana J.D., Meng X.G., Ting Y., Songwe F., Al Farga A., Mugisha S., Rwibasira P., Zhang Y.H., Zeng L.W. Anal. Biochem. 2020;600:13. doi: 10.1016/j.ab.2020.113762. [DOI] [PubMed] [Google Scholar]
- Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A., Garcia K.F., Barnes K.G., Chak B., Mondini A., Nogueira M.L., Isern S., Michael S.F., Lorenzana I., Yozwiak N.L., MacInnis B.L., Bosch I., Gehrke L., Zhang F., Sabeti P.C. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C., Wang R., Wu H., Zhang F., Wu J., Wang L. Anal. Chem. 2019;91:11362–11366. doi: 10.1021/acs.analchem.9b02554. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Qian C., Wu C., Wang Z., Wang D.C., Ye Z.Z., Ping J.F., Wu J., Ji F. Biosens. Bioelectron. 2020;157:112153. doi: 10.1016/j.bios.2020.112153. [DOI] [PubMed] [Google Scholar]
- Wu J., Liu J., Li S., Peng Z., Xiao Z., Wang X., Yan R., Luo J. Trav. Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101673. (in press), 101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.