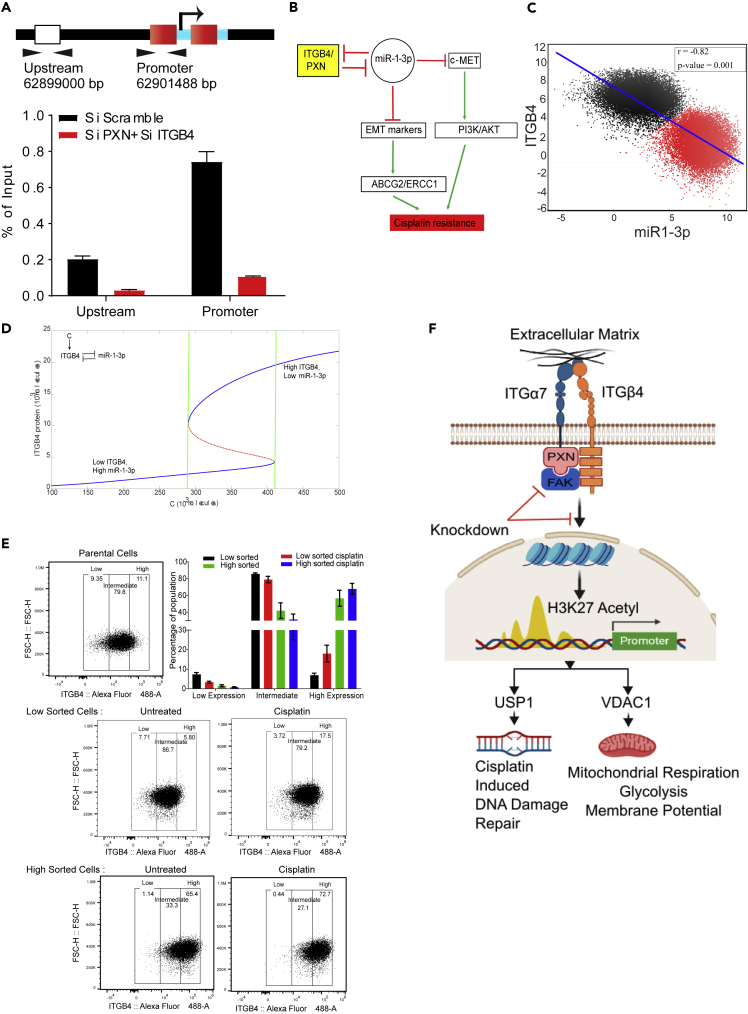
Figure 8.
Mathematical Modeling Suggests that Cisplatin Resistance Can Be Stochastic and Reversible
(A) Chromatin immunoprecipitation was performed with an acetylated H3K27 antibody 72 h after siRNA-mediated knockdown of PXN/ITGB4. With the knockdown, H3K27 acetylation at the promoter region of USP1 was greatly reduced compared with that of an upstream region, indicating the roles of ITGB4 and PXN in USP1 transcriptional activation.
(B) A double-negative feedback loop between ITGB4 and miR-1-3p leads to bistability.
(C) To test this mathematical model, RACIPE algorithm generated an ensemble (n = 100,000) with varying parameter sets then plotted to represent robust dynamical patterns. Results showed that ITGB4 and miR-1-3p exhibit bimodality: two distinct subpopulations of cells that are negatively correlated, reinforcing the previously described negative feedback loop.
(D) A mathematical model stimulating the dynamics of ITGB4 and miR-1-3p showed cisplatin resistance to be a reversible state. High ITGB4 and low miR-1-3p render cells to be resistant, whereas low ITGB4 and high miR-1-3p represents a more sensitive state.
(E) H2009 cells were stained with ITGB4 antibody conjugated to Alexa Fluor 488 and sorted based on gates set to high and low ~10% of ITGB4-expressing population using the FACSAria Fusion instrument. Sorted cells were subsequently cultured for 48 h and then treated with 1 μM cisplatin for 48 h. Then, using the Attune NxT Flow Cytometer, equal numbers of cells were stained again and analyzed to determine shifts in population between untreated and treated cells. Low sorted cells treated with cisplatin had a greater cell population that shifted toward higher ITGB4 expression. High sorted cells treated with cisplatin did not undergo significant changes in population compared with untreated.
(F) Schematic depicting the interaction between ITGB4 and PXN regulating downstream proteins USP1 and VDAC1 at the transcriptional level to coordinate cisplatin resistance. Data are represented as mean ± SD.