Abstract
Study rationale
The coexistence of KRAS and PIK3CA mutations in cells implies potential synergistic hyperactivation of the Ras/MAPK and PI3K/Akt oncogenic pathways. Therefore, it is desirable to investigate the concomitant mutations of KRAS and PIK3CA in colorectal cancer (CRC) samples and whether the concomitant mutations are associated with a poor prognosis in CRC patients.
Aim
To investigate the clinicpathological characteristics and prognostic value of concomitant mutations of KRAS and PIK3CA in CRC samples.
Methods
In this study, a total of 655 CRC patients from the Sixth Affiliated Hospital of Sun Yat-sen University were enrolled from January to December 2015. Sanger sequencing was applied to survey the mutational status of hotspot regions in the open reading frames (ORFs) of the KRAS and PIK3CA genes. Clinicpathological parameters were collected and analyzed. The Kaplan-Meier method and Cox regression model were applied to determine the correlation between the KRAS and PIK3CA mutation statuses and survival.
Results
We found that KRAS and PIK3CA bi-mutations were significantly associated with aggressive clinicpathological features. Among the studied CRC patients, those with either KRAS mutations (P = 0.004) or KRAS and PIK3CA bi-mutations (P = 0.033) had poor overall survival (OS). In the multivariable analysis, KRAS mutations in exons 3 and 4 but not exon 2 with concomitant PIK3CA mutations were associated with a high risk of death (univariate HR = 8.05; 95% CI, 1.926–33.64, P = 0.004; multivariate HR = 10.505; 95% CI, 2.304–47.905, P = 0.002).
Conclusion
The concomitant mutation statuses of KRAS and PIK3CA should be considered when the prognostic value of gene mutations is consulted in CRC patients.
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide, including China, with morbidity and mortality rates ranking in the top five among all tumors [1]. It has generally been accepted that the occurrence and development of tumors are related to the abnormal activation of many signaling pathways, among which the classic pathways include the mitogen-activated protein kinase (MAPK) and phosphatidyl-inositol-3 kinase (PI3K) signaling pathways [2].
Rat sarcoma viral oncogene homolog (RAS) genes, including v-Ki-ras2-Kirsten rat sarcoma viral oncogene homolog (KRAS) and neuroblastoma rat sarcoma viral oncogene homolog (NRAS), were the first confirmed human proto-oncogenes that can transform into oncogenes. KRAS is the most frequently mutated RAS member in most cancers and is altered at high frequencies (30–50%) in CRC patients [3]. Most KRAS mutations are detected in codon 12 or 13 of exon 2 and account for nearly 90% of all mutation types; other mutations in codon 59 or 61 of exon 3 and codon 117 or 146 of exon 4 occur less frequently [4]. KRAS mutation often leads to the guanosine triphosphate (GTP)-bound form of the coded protein. This then results in the persistent activation of downstream signaling pathways such as the MAPK and PI3K pathways. Although KRAS has been validated as a molecular biomarker for anti- epidermal growth factor receptor (EGFR) therapy [5], the prognostic value of KRAS mutation is still controversial.
Recently, an analysis from the National Cancer Database involving 19,877 nonmetastatic colon cancer patients showed that KRAS-mutated tumors were more frequently observed in right sided colon and late-stage tumors and associated with a poor prognosis in stage III patients [6]. A similar result was obtained in a cohort of 2720 patients by Sinicrope et al. [7], the 5-year disease-free survival (DFS) rate was 70.7% for patients with wild-type (WT) KRAS and 61% for patients with mutant KRAS. In metastatic patients, KRAS has been reported to be associated with poor recurrence-free survival and overall survival (OS) after curative resection [8]. Indeed, Ye et al. [9]; studied a large cohort of 1190 Chinese CRC patients and found no significant difference in OS between KRAS WT and mutant patients.
Several studies have revealed a strong association of KRAS and Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) gene mutations in CRC [10,11]. PIK3CA is involved in the PI3K/Akt signaling pathway and is associated with high mutation rates in CRC (10–20%), second only to KRAS [12], and its somatic activating mutation plays an important role in tumorigenesis [13]. In 2007, Kato et al. [14]; found that the OS time for stage II and III CRC patients with mutant PIK3CA was significantly shorter than that for CRC patients with WT PIK3CA (P = 0.043, HR = 2.478). Successive studies have also found that PIK3CA mutation is related to postoperative recurrence, distant lung or liver metastases, chemotherapy resistance and other adverse prognostic events in CRC patients [[15], [16], [17], [18], [19], [20], [21]]. Multiple studies have found that stage IV CRC patients with PIK3CA mutation are resistant to EGFR monoclonal antibody therapy [[22], [23], [24], [25]].
Nevertheless, some experts have noted that PIK3CA is a favorable biomarker for the prognosis of stage I-III patients [26]. CRC patients with a somatic mutation in PIK3CA may benefit from aspirin administration [27]. Zuo et al. [28]; showed that somatic PIK3CA mutations might be related to radiotherapy sensitivity, suggesting that CRC patients with liver metastasis who are resistant to EGFR monoclonal antibodies could benefit from artery radiotherapy embolization therapy. Therefore, the prognostic value of PIK3CA mutation in CRC remains controversial.
At face value, KRAS and PIK3CA bi-mutations may represent the co-activation of MAPK and PI3K signaling pathways. Therefore, it is reasonable to conjecture that the coexistence of KRAS and PIK3CA mutations may have synergistic or additive effects on survival in CRC patients. Some scholars have even noted that the prognostic and predictive value of PIK3CA may actually depend on the mutational status of KRAS [[29], [30], [31]]. Therefore, whether the concomitant mutations of KRAS and PIK3CA are potential markers for a poor prognosis in CRC patients requires further exploration.
Materials and methods
Patients
In total, 655 CRC patients who underwent surgical resection at the Sixth Affiliated Hospital of Sun Yat-sen University (SYSU) between January and December 2015 and met the criteria below were included in this retrospective study. All patients signed an informed consent form, which was approved by the Institutional Review Board of the hospital.
The inclusion criteria were as follows: (1) the resected CRC tumor was histologically confirmed; (2) case data and follow-up records were complete; and (3) formalin-fixed, paraffin-embedded specimens were available. The exclusion criterion was as follows: accompanied by other types of cancer or severe diseases such as infection or organ dysfunction.
Detection of KRAS and PIK3CA mutations
Sanger sequencing was performed in the Molecular Diagnostic Laboratory of the Sixth Affiliated Hospital of Sun Yat-sen University. KRAS and PIK3CA mutation testing was performed with Sanger sequencing and HRM method developed by the Lab. And the detail procedure of the mutation testing can refer to previous studies [32,33].
Statistical analysis
The correlation between KRAS and PIK3CA mutation statuses and clinicpathological characteristics was evaluated using a chi square test (or Fisher's exact test) for categorical data. OS was defined as the period from the date of surgery to death from any cause. DFS was defined as the period from the date of surgery to tumor recurrence or death. The Kaplan-Meier method was performed to compare OS and DFS between groups. Univariate and multivariate Cox proportional hazards models were used to explore the associations of patient characteristics and gene mutations with OS and DFS. Two-sided p-values are reported, and, in general, P-values <0.05 were considered statistically significant. Analyses were performed using SPSS software version 20.0 (SPSS Inc., Chicago, IL, USA).
Transcriptomic analysis
We also looked at the TCGA CRC portal with attention for cases with gene mutation and expression data matched/available (233 cases). The mutational and expression data and related clinical records were downloaded. Differentially expressed genes among different groups were identified by using the R language limma package (version 3.6; https://www.r-project.org/) (P < 0.05 and |log 2 FC| > 1). GO function enrichment analysis and KEGG pathway analysis were performed using the clusterProfiler package.
Results
A total of 655 patients were included in this study, and their clinicpathological characteristics are shown in Supplementary Table 1. The CRC incidence rates were higher in males than in females (59.5% vs 40.5%, respectively). Patients with CRC had a similar age distribution at the time of surgery. Abnormal levels of tumor markers were observed in less than half of CRC patients. The rectum was the most common primary tumor site, followed by the left sided colon and right sided colon (47.5%, 30.7%, and 19.7%, respectively). Nearly half of the CRC patients had advanced tumors (48.7%, 319/655). In total, 20.5%, 60.9%, and 18.5% of tumors were graded as 1, 2 and 3, respectively. A small proportion of patients overexpressed the HER2 oncogene (36.5%, 234/641). High KI-67 expression was observed in 26.6% (174/645) of patients with CRC. Perineural invasion (6.6%, 43/649) and lymph vascular invasion (8.2%, 53/647) were rarely observed in this cohort. Deficient mismatch repair (dMMR) was observed in 6.4% of CRC patients (42/651). The clinicpathological characteristics of this TCGA cohort are summarized in Supplementary Table 2.
Gene mutations
Of the 655 patients, the mutation rate of KRAS was 46.6% (305 of 655; exon 2 (n = 259) and exon 3 or 4 (n = 46)), and that of PIK3CA was 12.8% (84 of 655; exon 9 (n = 61) and exon 20 (n = 22)) (Supplementary Table 3). KRAS and PIK3CA concomitant mutations were commonly observed in PIK3CA-mutated tumors (55/84, 65.5%) (Table 1). Only one patient harbored a PIK3CA mutation in both exons 9 and 20 concomitant with a KRAS mutation in exon 4. We also performed similar analysis on the TCGA CRC dataset, 223 cases of patients with KRAS or PIK3CA mutation information available. The mutation rate of KRAS and PIK3CA were 43% (97 of 223; exon 2 (n = 79), exon 3 or 4 (n = 17)) and 15.2% (34 of 223; exon 10(n = 13), exon 21(n = 6) and other exons (n = 15)), respectively. KRAS and PIK3CA concomitant mutations were commonly observed in PIK3CA-mutated tumors (25/34, 73.5%). Five cases were detected more than one exon mutations in PIK3CA, and all of them were concurrent mutations with KRAS. However, we could not find the coexistence of PIK3CA mutations in exon 9 or 20 and KRAS mutations in exon 3 or 4 (Supplementary Tables 4, 5).
Table 1.
Distribution of KRAS and PIK3CA concomitant mutations among 655 CRC patients.
| KRAS |
Exon 2 |
Exon 3 |
Exon 4 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIK3CA | p.G12S | p.G12D | p.G12A | p.G12V | p.G12C | p.G12P | p.G13D | p.G13C | p.A59T | p.Q61K | p.Q61H | p.A146T | |
| Exon 9 | p.E542K | 6 | 1 | 1 | 1 | ||||||||
| p.E545K | 1 | 11 | 4 | 1 | 6 | 1 | 1 | 1 | |||||
| p.E545D | 1 | ||||||||||||
| p.E545A | 1 | 1 | 1 | ||||||||||
| p.E545G | 1 | ||||||||||||
| Exon 20 | p.H1047L | 1 | 5 | 1 | 2 | 2 | 1 | ||||||
| p.H1047R | 1 | 1 | |||||||||||
| p.E545G, p.H1047Y | 1 | ||||||||||||
Correlations with clinical characteristics
As showed in Table 2, there was a significant difference categorized either by sex, age, primary tumor site, levels of CA-199 and CA125, microsatellite instability (MSI) status, TNM stage, and tumor grade (all with P < 0.05). Tumors with KRAS and PIK3CA bi-mutations tended to be located in a proximal location (P<0.001), poorly differentiated (P = 0.019), and have elevated CA-199 (P = 0.001) and CA125 levels (P = 0.033). CRC patients were more likely to be diagnosed in the late stage than in the early stage (P = 0.002) at the initial diagnosis. Compared with individual KRAS mutations, additional PIK3CA mutations were associated with more advanced tumors (P = 0.008). There were more cases with dMMR in the individual PIK3CA mutation group than in the bi-mutation group (P = 0.032). However, we found no significant association between mutation type and clinicpathological characteristics in TCGA dataset (Supplementary Table 6).
Table 2.
Correlations between gene mutation types and clinicpathological characteristics.
|
KRAS and PIK3CA wild-type |
KRAS mutations alone |
PIK3CA mutations alone |
KRAS and PIK3CA bi-mutations |
P value |
P value |
P value |
P value |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | (bi-mutations vs wild-type) | (bi-mutations vs KRAS mutations alone) | (bi-mutations vs PIK3CA mutations alone) | ||
| Sex | ||||||||||||
| Female | 113 | 35.20 | 112 | 44.80 | 17 | 58.60 | 23 | 41.80 | 0.022⁎ | 0.345 | 0.687 | 0.143 |
| Male | 208 | 64.80 | 138 | 55.20 | 12 | 41.40 | 32 | 58.20 | ||||
| Age, years | ||||||||||||
| <60 years | 175 | 54.50 | 109 | 43.60 | 14 | 48.30 | 23 | 41.80 | 0.047⁎ | 0.081 | 0.809 | 0.571 |
| ≥60 years | 146 | 45.50 | 141 | 56.40 | 15 | 51.70 | 32 | 58.20 | ||||
| CEA level | ||||||||||||
| 0–5 ng/ml | 197 | 62.30 | 128 | 53.10 | 18 | 62.10 | 33 | 60.00 | 0.171 | 0.741 | 0.355 | 0.854 |
| >5 ng/ml | 119 | 37.70 | 113 | 46.90 | 11 | 37.90 | 22 | 40.00 | ||||
| CA-199 level | ||||||||||||
| 0–37 ng/ml | 285 | 89.90 | 192 | 79 | 25 | 86.20 | 41 | 74.50 | 0.001⁎ | 0.001⁎ | 0.469 | 0.216 |
| > 37 ng/ml | 32 | 10.10 | 51 | 21.00 | 4 | 13.80 | 14 | 25.50 | ||||
| CA-125 level | ||||||||||||
| 0–35 ng/ml | 296 | 93.70 | 220 | 90.50 | 22 | 75.90 | 47 | 85.50 | 0.005⁎ | 0.033⁎ | 0.265 | 0.275 |
| >35 ng/ml | 20 | 6.30 | 23 | 9.50 | 7 | 24.10 | 8 | 14.50 | ||||
| HER2 | ||||||||||||
| Positive | 122 | 39.00 | 91 | 37.00 | 7 | 24.10 | 14 | 26.40 | 0.165 | 0.08 | 0.143 | 0.821 |
| Negative | 191 | 61.00 | 155 | 63.00 | 22 | 75.90 | 39 | 73.60 | ||||
| KI67 | ||||||||||||
| Positive | 99 | 31.30 | 60 | 24.40 | 5 | 17.20 | 10 | 18.50 | 0.065 | 0.056 | 0.356 | 0.885 |
| Negative | 217 | 68.70 | 186 | 75.60 | 24 | 82.80 | 44 | 81.50 | ||||
| Tumor location | ||||||||||||
| Proximal colon | 43 | 13.60 | 52 | 21.50 | 10 | 34.50 | 24 | 44.40 | <0.001⁎ | <0.001⁎ | 0.002⁎ | 0.013⁎ |
| Distal colon | 117 | 37.00 | 60 | 24.80 | 14 | 48.30 | 10 | 18.50 | ||||
| Rectum | 156 | 49.40 | 130 | 53.70 | 5 | 17.20 | 20 | 37.00 | ||||
| MSI status | ||||||||||||
| MSI-L/MSI-H | 20 | 6.30 | 13 | 5.20 | 6 | 20.70 | 3 | 5.50 | 0.015⁎ | 0.812 | 0.944 | 0.032⁎ |
| MSS | 298 | 93.70 | 236 | 94.80 | 23 | 79.30 | 52 | 94.50 | ||||
| T | ||||||||||||
| Tis-T1 | 20 | 6.20 | 10 | 4.00 | 0 | 0.00 | 2 | 3.60 | 0.055 | 0.124 | 0.518 | 0.685 |
| T2 | 40 | 12.50 | 31 | 12.40 | 1 | 3.40 | 3 | 5.50 | ||||
| T3 | 242 | 75.40 | 178 | 72.10 | 23 | 79.30 | 43 | 78.20 | ||||
| T4 | 19 | 5.90 | 31 | 12.40 | 5 | 17.20 | 7 | 12.70 | ||||
| N | ||||||||||||
| N0 | 178 | 55.50 | 136 | 54.40 | 17 | 58.60 | 32 | 58.20 | 0.353 | 0.656 | 0.175 | 0.934 |
| N1 | 94 | 29.30 | 86 | 34.40 | 6 | 20.70 | 13 | 23.60 | ||||
| M | ||||||||||||
| M0 | 271 | 84.16 | 212 | 84.80 | 23 | 79.30 | 37 | 67.30 | 0.012⁎ | 0.002⁎ | 0.002⁎ | 0.247 |
| M1 | 50 | 15.53 | 38 | 15.20 | 6 | 20.70 | 18 | 32.70 | ||||
| TNM staging | ||||||||||||
| I | 39 | 12.10 | 28 | 11.20 | 0 | 0.00 | 4 | 7.30 | 0.023⁎ | 0.007⁎ | 0.008⁎ | 0.246 |
| II | 126 | 39.30 | 100 | 40.00 | 16 | 55.20 | 23 | 41.80 | ||||
| III | 106 | 33.00 | 84 | 33.60 | 7 | 24.10 | 10 | 18.20 | ||||
| IV | 50 | 15.60 | 38 | 15.20 | 6 | 20.70 | 18 | 32.70 | ||||
| Tumor grade | ||||||||||||
| Grade 3 | 46 | 14.40 | 49 | 19.70 | 10 | 34.50 | 16 | 29.10 | 0.009⁎ | 0.019⁎ | 0.074 | 0.871 |
| Grade 2 | 200 | 62.50 | 159 | 63.90 | 13 | 44.80 | 26 | 47.30 | ||||
| Grade 1 | 74 | 23.10 | 41 | 16.50 | 6 | 20.70 | 13 | 23.60 | ||||
| Perineural invasion | ||||||||||||
| No | 293 | 92.70 | 234 | 94.40 | 26 | 89.70 | 52 | 94.50 | 0.712 | 0.625 | 0.956 | 0.408 |
| Yes | 23 | 7.30 | 14 | 5.60 | 3 | 10.30 | 3 | 5.50 | ||||
| Lymphovascular invasion | ||||||||||||
| No | 290 | 92.40 | 229 | 92.00 | 25 | 86.20 | 50 | 90.90 | 0.705 | 0.713 | 0.796 | 0.508 |
| Yes | 24 | 7.60 | 20 | 8.00 | 4 | 13.80 | 5 | 9.10 | ||||
P-values≤0.05.
Survival analysis
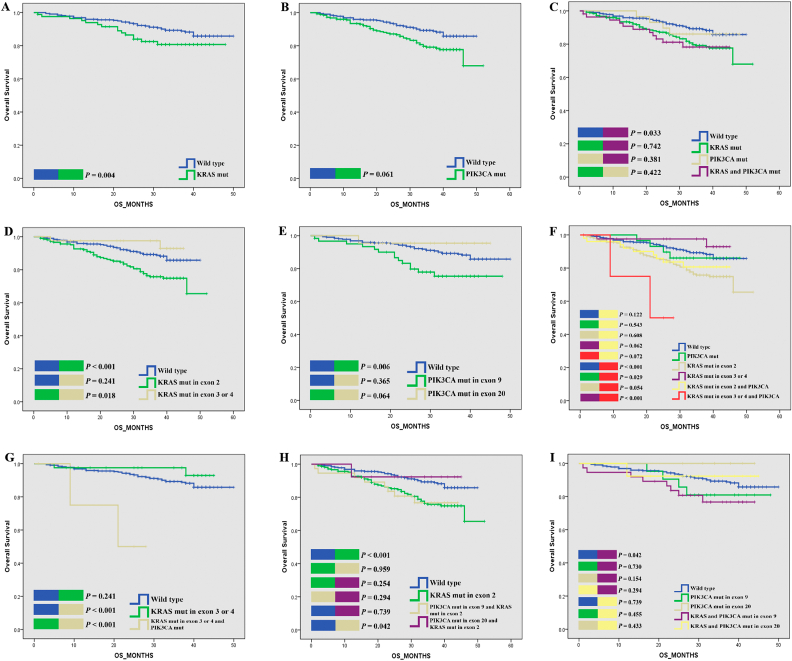
To clarify whether KRAS/PIK3CA gene mutations are associated with longer or shorter OS and DFS, we used the Kaplan-Meier method to compare OS and DFS between patients with KRAS or PIK3CA mutant and WT tumors, and found that patients with mutant KRAS experienced shorter OS than patients with WT KRAS (Log-rank P = 0.004) (Fig. 1A). Furthermore, patients with KRAS mutations in exon 2 experienced significantly poor OS than KRAS mutations in exon 3 and 4 (KRAS exon 2 vs. KRAS exon 3 and 4; Log-rank P = 0.018) (Fig. 1D). No significant difference in OS was found between patients with mutant PIK3CA and those with WT PIK3CA (Log-rank P = 0.061) (Fig. 1B). However, subgroup analysis showed that patients with PIK3CA mutations in exon 9 (vs. WT, Log-rank P = 0.006) (Fig. 1E) experienced worse OS than patients with PIK3CA mutations in exon 20 (vs. WT, Log-rank P = 0.365). The Kaplan–Meier plots of TCGA dataset shown that neither KRAS or PIK3CA mutations had significant impact on OS status (Supplementary Fig. 1A, B, D).
Fig. 1.
Kaplan-Meier plots of OS for CRC patients according to gene mutation status.
Then, the group with KRAS and PIK3CA bi-mutations was examined. As shown in Fig. 1H, KRAS exon 2 group has poorer OS (vs. WT, Log-rank P < 0.001), with a similar trend like the PIK3CA exon 9 and KRAS exon 2 group (vs. WT, Log-rank P = 0.042), but not the PIK3CA exon 20 and KRAS exon 2 group (vs. WT Log-rank P = 0.739). Fig. 1I shown that although PIK3CA exon 9 and KRAS exon 2 has poorer OS, but there have no statistically significant difference than other groups including the PIK3CA exon 9, the PIK3CA exon 20 group and the PIK3CA exon 20 and KRAS exon 2 group (Log-rank P > 0.05). As shown in Fig. 1C and Supplementary Table 7, we observed a trend toward worse OS in patients with concomitant mutations, but that trend was not significant in the multivariate analysis (Log-rank P = 0.033, univariate HR = 2.078; 95% CI, 1.059–4.079, P = 0.034; multivariate HR = 1.281; 95% CI, 0.609–2.692, P = 0.514). Only if the PIK3CA mutation coexisted with the KRAS mutation in exons 3 and 4 did patients experience much shorter OS than other patients (Log-rank P < 0.001, univariate HR = 8.05; 95% CI, 1.926–33.64, P = 0.004; multivariate HR = 10.505; 95% CI, 2.304–47.905, P = 0.002) (Fig. 1F, G,Table 3). The Kaplan–Meier plots of TCGA dataset shown that, the KRAS and PIK3CA bi-mutation group had no statistical difference on OS when compare to other groups (Supplementary Fig. 1C). Whereas, the Cox regression model shown that in the multivariate but not univariate analysis, the KRAS and PIK3CA bi-mutation group had poor OS than WT (univariate HR = 1.204; 95% CI, 0.517, 2.805, P = 0.667; multivariate HR = 3.010;95% CI, 0.991, 9.149, P = 0.052) (Supplementary Table 8). When referring to specific KRAS mutation types, the PIK3CA and KRAS exon 2 group have poorer OS than the KRAS exon group (Log-rank P = 0.041, Supplementary Fig. 1E); when compared to WT, the PIK3CA and KRAS exon 2 group have a poorer OS (univariate HR = 1.575; 95% CI, 0.675,3.675, P = 0.293; multivariate HR = 3.864;95% CI, 1.236,12.080, P = 0.020) (Supplementary Table 9).
Table 3.
Univariate and multivariate analyses of different prognostic parameters regarding to OS of the CRC patients.
| Univariate analysis |
Multivariate analysis |
|||
|---|---|---|---|---|
| Parameters | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (<60 vs. ≥ 60 years) | 0.690 (0.464,1.027) | 0.068 | ||
| Gender (Female vs. Male) | 0.911 (0.609,1.363) | 0.649 | ||
| Histological grade | 0.610 (0.446,0.834) | 0.002⁎ | 0.754 (0.517,1.100) | 0.142 |
| CEA level (>5 ng/ml vs. ≤5 ng/ml) | 3.512 (2.280,5.408) | <0.001⁎ | 1.510 (0.914,2.496) | 0.108 |
| CA19–9 level (>37 ng/ml vs. ≤37 ng/ml) | 5.137 (3.440,7.672) | <0.001⁎ | 2.130 (1.285,3.532) | 0.003⁎ |
| CA12–5 level (>35 ng/ml vs. ≤35 ng/ml) | 3.251 (2.006,5.271) | <0.001⁎ | 1.401 (0.783,2.505) | 0.256 |
| Tumor location | 0.003⁎ | 0.405 | ||
| Proximal colon | 2.281 (1.359,3.830) | 0.002⁎ | 1.203 (0.663,2.183) | 0.544 |
| Distal colon | 1.963 (1.230,3.133) | 0.005⁎ | 1.419 (0.852,2.365) | 0.179 |
| Rectum | 1 | 1 | ||
| TNM stage | 3.731 (2.864,4.861) | <0.001⁎ | 2.153 (1.459,3.178) | <0.001⁎ |
| HER2 (Negative vs Positive) | 1.31 (0.854,2.008) | 0.216 | ||
| KI-67 (Negative vs Positive) | 1.485 (0.91,2.424) | 0.114 | ||
| MSI status (dMMR vs pMMR) | 0.286 (0.07,1.158) | 0.079 | ||
| Perineural invasion (Yes vs no) | 3.109 (1.864,5.184) | <0.001⁎ | 0.974 (0.485,1.958) | 0.941 |
| Lymphovascular invasion (Yes vs no) | 2.650 (1.504,4.668) | 0.001⁎ | 1.952 (1.068,3.565) | 0.03⁎ |
| Chemotherapy or radiotherapy (Yes vs No) | 1.426 (0.953,2.133) | 0.085 | 0.476 (0.294,0.771) | 0.003⁎ |
| Radical Surgery (No vs Yes) | 11.592(7.804,17.217) | <0.001⁎ | 3.526 (1.891,6.576) | <0.001⁎ |
| Mutations status | 0.001⁎ | 0.001⁎ | ||
| KRAS/PIK3CA wild-type | 1 | 1 | ||
| PIK3CA mut alone | 1.246 (0.444,3.496) | 0.677 | 0.660 (0.219,1.994) | 0.462 |
| KRAS mutation in exon 2 | 2.147 (1.395,3.303) | 0.001⁎ | 2.185 (1.338,3.569) | 0.002⁎ |
| KRAS mutation in exon 3 or 4 | 0.451 (0.109,1.870) | 0.272 | 0.868 (0.206,3.669) | 0.848 |
| Coexistence of PIK3CA mutant and KRAS mutation in exon 2 | 1.786 (0.861–3.704) | 0.119 | 1.080 (0.486–2.398) | 0.850 |
| Coexistence of PIK3CA mutant and KRAS mutation in exon 3 or 4 | 8.050 (1.926,33.644) | 0.004⁎ | 10.505 (2.304–47.905) | 0.002⁎ |
Hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated using univariate or multivariate Cox proportional hazards regression in SPSS 20.0. P-values were calculated using univariate or multivariate Cox proportional hazards regression in SPSS 20.0. P-values <0.05 were considered to indicate statistical significance.
P < 0.05.
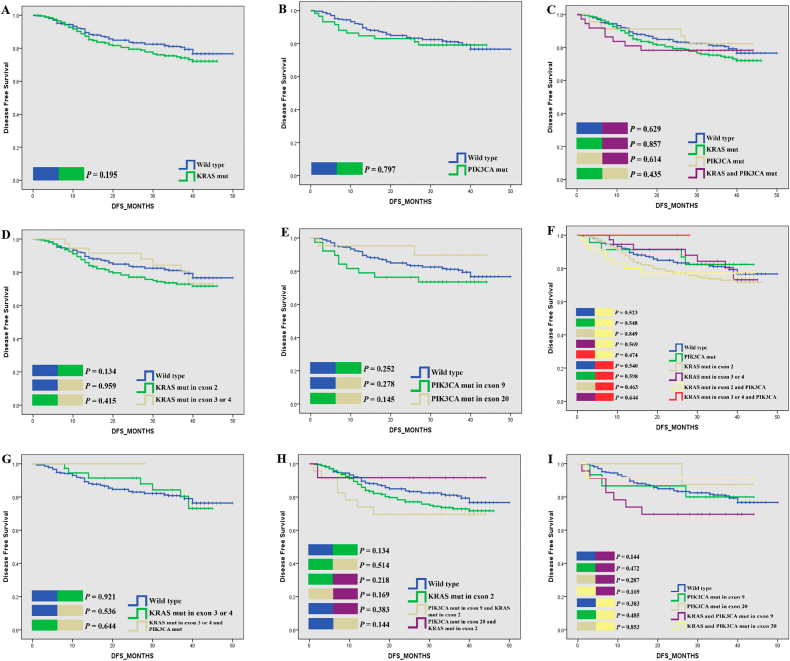
With the same setting of groups and analysis procedures, we also compared the association between mutational status and DFS. However, no significant difference in DFS was found between these groups, although the KRAS mutations group, especially the KRAS exon 2 group have poorer DFS than WT, but no significant different were found in further analysis (Fig. 2, Supplementary Fig. 2).
Fig. 2.
Kaplan-Meier plots of DFS for CRC patients according to gene mutation status.
Transcriptomic analysis
We used downloaded data from TCGA portal with gene mutation information and expression data matched for the transcriptomic analysis. Differentially expressed genes (DEGs) between various groups were identified first as described in methods. Then the list of DEGs were subjected to enrichment analysis in terms of molecular functions, biological processes, and cellular components, or KEGG pathways (Supplementary Figs. 3–5, Supplementary Table 10). We speculate that different subtype might lead to different tumor metabolism and this phenomenon should be considered in stratification of chemotherapy and future drug development accordingly. Subgroup analysis were also performed (Supplementary Figs. 6–12). Given the small size of cohort and detailed clinical information partially missed, solid conclusion is yet to be explored and open to question.
Discussion
Hyperactivation of the MAPK and PI3K signaling pathways can lead to uncontrolled cell proliferation and apoptosis, further transformation of carcinogenesis and invasion and metastasis [2]. In this study, a strong association of KRAS and PIK3CA mutations was revealed. Simultaneous mutations in PIK3CA exon 9 and KRAS exon 2 were the most common (37/55,67.2%), consistent with the findings by Li et al. [15]. Compared with WT tumors, tumors with KRAS and PIK3CA bi-mutations were significantly associated with aggressive clinicpathological features. For example, increased proportions of proximal colon cancer, were prone to metastasize to distal organs at initial surgery, and high-grade tumors tended to have elevated CA19-9 and CA125 levels. Compared to CRC patients with individual KRAS mutations, those with bi-mutations had more advanced tumors that were more frequently located in the right sided colon than in the rectum and left sided colon (44.4% vs 21.5%, 37% vs 53.7%, and 18.5% vs 24.8%, respectively, P = 0.002). The major differences observed between single PIK3CA mutations and KRAS and PIK3CA bi-mutations were the MSI status and tumor location, consistent with previous studies in which the PIK3CA mutation was reported to be closely related to the distal colon and dMMR status [19]. We also try to validated our findings in CRC cohort from TCGA. However, we found no significant association between mutation type and clinicpathological characteristics. Although please keep in mind that the small sample size might give rise to statistical bias (Supplementary Tables 2, 6).
In our study, we found that KRAS mutations are consistently associated with poor prognosis in both univariate and multivariate analysis (univariate HR = 2.147; 95% CI, 1.395,3.303, P = 0.001; multivariate HR = 2.185; 95% CI, 1.338,3.569, P = 0.002) (Table 3). Among the specific mutations in KRAS, those in exon 2 but not exons 3 and 4 showed a significant association with OS (Log-rank P<0.001) (Fig. 1D). Regarding the potential co-selection of KRAS and PIK3CA, we observed a trend toward worse OS in patients with concomitant mutations, but not indicated in multivariate analysis (univariate HR = 2.078; 95% CI, 1.059–4.079, P = 0.034, multivariate HR = 1.281; 95% CI, 0.609–2.692, P = 0.514) (Supplementary Table 7). Therefore, we initially speculated that the prognosis of PIK3CA and KRAS concomitant mutations may depend on the KRAS mutational status. However, further study shown that patients with KRAS mutation in exon 3 and 4 as well as PIK3CA mutation have much shorter OS than other patients (univariate HR = 8.05; 95% CI, 1.926–33.64, P = 0.004; multivariate HR = 10.505; 95% CI, 2.304–47.905, P = 0.002,Table 3). Not only that, multivariate analysis shown the HR of this particular group are much higher than the KRAS exon 2 mutations group (univariate HR = 2.147;95% CI,1.395–3.303, P = 0.001; multivariate HR =2.185;95% CI,1.338–3.569, P = 0.002), also much higher than the PIK3CA and KRAS exon 2 group (univariate HR = 1.786;95% CI, 0.861–3.704, P = 0.119; multivariate HR =1.080; 95% CI, 0.486–2.398, P = 0.850). We performed similar analysis on the CRC cohort from TCGA, the results were consistent with our conclusion that the bi-mutations have poorer OS, but only refer to KRAS mutations in exon 2 (univariate HR = 1.575; 95% CI, 0.675–3.675, P = 0.293, multivariate HR = 3.864, 95% CI, 1.236–12.080, P = 0.020) (Supplementary Tables 8, 9). In summary, the concomitant mutations did have synergistic effect on survival status in CRC patients. We also found that the KRAS and PIK3CA mutation status may influence OS but not DFS (Fig. 2). We speculate that this observation is mainly because we performed radical surgery early after the initial diagnosis in stage I-III CRC patients.
Currently, many studies have suggested that the KRAS and PIK3CA mutation status is significantly associated with the chemotherapy response. When compare to others, the bi-mutations tumor may lead to distinct tumor metabolism and tumor immune response which hints that the corresponding therapy need in-depth study (Supplementary Figs. 3–12, Supplementary Table 10). EGFR monoclonal antibody therapy combined with the FOLFIRI or FOLFOX regimen has shown significant advantages in selected patients with stage IV CRC. However, many large clinical trials have found that CRC tumors with KRAS or PIK3CA gene mutations do not respond to EGFR monoclonal antibody therapy [34,35]. Jhawer et al. [36]; found that cetuximab had no effect on cell lines with KRAS exon 2 and PIK3CA exon 20 mutations but WT or individual KRAS/PIK3CA mutations. MEK inhibitors are potential drugs for the treatment of CRC; however, the efficacy of a single therapy on CRC tumors with KRAS mutations varies greatly [37]. Studies have shown that PIK3CA mutations can reduce the sensitivity of KRAS-mutant CRC cells to MEK inhibitors, but MEK inhibitors combined with PI3K signaling pathway inhibitors can improve the treatment effect [38]. However, due to the high cost of targeted therapy drugs, fluorouracil-based chemotherapy is still the major treatment option for advanced CRC treatment in China. In this study, among the 112 metastatic CRC patients, 87 chose postoperative chemotherapy, and a few were treated with EGFR or VEGF monoclonal antibody therapy after surgery. Therefore, the prognostic impact of targeting drugs on this small cohort could not be adequately evaluated.
In conclusion, our study suggests that KRAS and PIK3CA bi-mutations are associated with more aggressive clinicpathological features and should be regarded as new addition of molecular stratification, especially when referring to specific KRAS mutation types. Longer follow-up times and increased sample sizes are required for more stringent findings, and clinical trials are needed to validate the value of these mutations in the comprehensive management of CRC patients.
CRediT authorship contribution statement
Qianxin Luo: Conceptualization, data collection and analysis, manuscript draft preparation.
Dianke Chen: Conceptualization, methodology guidance.
Xinjuan Fan: Resources.
Xinhui Fu: Resources.
Tenghui Ma: Manuscript editing.
Daici Chen: Conceptualization, supervision, manuscript preparation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
Dr. Lei Wang was one of the pioneers for the Sixth Affiliated Hospital of Sun Yat-sen University. We were deeply sorrowful for the loss of him. We will move on with what he had taught and inspired us. We also sincerely thank the Guangdong Research Institute of Gastroenterology, Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Diseases and Department of Colorectal Surgery, and the Sixth Affiliated Hospital, Sun Yat-sen University.
Ethical approval
Our study was approved by the Institutional Ethics Committee of Sixth Affiliated Hospital, Sun Yat-sen University. The related ethical approval code is: 2018–071.
Funding statement
This work was supported by the National Natural Science Foundation of China, No. 31970703; the Natural Science Foundation of Guangdong Province No. 2017A030313805.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100874.
Appendix A. Supplementary data
Supplementary material
References
- 1.Chen W., Zheng R., Baade P.D. Cancer statistics in China, 2015[J] CA Cancer J. Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Papadatos-Pastos D., Rabbie R., Ross P. The role of the PI3K pathway in colorectal cancer[J] Crit Rev Oncol Hematol. 2015;94(1):18–30. doi: 10.1016/j.critrevonc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Bonnot P.E., Passot G. RAS mutation: site of disease and recurrence pattern in colorectal cancer[J] Chin Clin Oncol. 2019;8(5):55. doi: 10.21037/cco.2019.08.11. [DOI] [PubMed] [Google Scholar]
- 4.Tan C., Du X. KRAS mutation testing in metastatic colorectal cancer[J] World J. Gastroenterol. 2012;18(37):5171–5180. doi: 10.3748/wjg.v18.i37.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinu D., Dobre M., Panaitescu E. Prognostic significance of KRAS gene mutations in colorectal cancer--preliminary study[J] J Med Life. 2014;7(4):581–587. [PMC free article] [PubMed] [Google Scholar]
- 6.Scott A., Goffredo P., Ginader T. The impact of KRAS mutation on the presentation and prognosis of non-metastatic colon cancer: An analysis from the National Cancer Database[J] J. Gastrointest. Surg. 2020 doi: 10.1007/s11605-020-04543-4. [DOI] [PubMed] [Google Scholar]
- 7.Sinicrope F.A., Shi Q., Smyrk T.C. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes[J] Gastroenterology. 2015;148(1):88–99. doi: 10.1053/j.gastro.2014.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vauthey J.N., Zimmitti G., Kopetz S.E. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases[J] Ann. Surg. 2013;258(4):619–626. doi: 10.1097/SLA.0b013e3182a5025a. 626-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye Z.L., Qiu M.Z., Tang T. Gene mutation profiling in Chinese colorectal cancer patients and its association with clinicopathological characteristics and prognosis[J] Cancer Med. 2020;9(2):745–756. doi: 10.1002/cam4.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Roock W., Claes B., Bernasconi D. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis[J] Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 11.Susanti S., Fadhil W., Murtaza S. Positive association of PIK3CA mutation with KRAS mutation but not BRAF mutation in colorectal cancer suggests co-selection is gene specific but not pathway specific[J] J. Clin. Pathol. 2019;72(3):263–264. doi: 10.1136/jclinpath-2018-205483. [DOI] [PubMed] [Google Scholar]
- 12.Cathomas G. PIK3CA in colorectal Cancer[J] Front. Oncol. 2014;4:35. doi: 10.3389/fonc.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samuels Y., Diaz L.J., Schmidt-Kittler O. Mutant PIK3CA promotes cell growth and invasion of human cancer cells[J] Cancer Cell. 2005;7(6):561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Kato S., Iida S., Higuchi T. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer[J] Int. J. Cancer. 2007;121(8):1771–1778. doi: 10.1002/ijc.22890. [DOI] [PubMed] [Google Scholar]
- 15.Li H.T., Lu Y.Y., An Y.X. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer[J] Oncol. Rep. 2011;25(6):1691–1697. doi: 10.3892/or.2011.1217. [DOI] [PubMed] [Google Scholar]
- 16.Mao C., Yang Z.Y., Hu X.F. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis[J] Ann. Oncol. 2012;23(6):1518–1525. doi: 10.1093/annonc/mdr464. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y., Han X., Wang J. Prognostic impact of mutation profiling in patients with stage II and III colon cancer[J] Sci. Rep. 2016;6:24310. doi: 10.1038/srep24310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo A.L., Borger D.R., Szymonifka J. Mutational analysis and clinical correlation of metastatic colorectal cancer[J] Cancer. 2014;120(10):1482–1490. doi: 10.1002/cncr.28599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosty C., Young J.P., Walsh M.D. PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival[J] PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0065479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang M., Shen X.J., Kim S. Somatic gene mutations in African Americans may predict worse outcomes in colorectal cancer[J] Cancer Biomark. 2013;13(5):359–366. doi: 10.3233/CBM-130366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang L., Wei Y., Ren L. [A study of the relationship between the mutation of PIK3CA, PTEN and the occurrence of liver metastasis of colorectal cancer: survival analysis][J] Zhonghua Wai Ke Za Zhi. 2012;50(11):1007–1010. [PubMed] [Google Scholar]
- 22.Sartore-Bianchi A., Martini M., Molinari F. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies[J] Cancer Res. 2009;69(5):1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 23.Therkildsen C., Bergmann T.K., Henrichsen-Schnack T. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis[J] Acta Oncol. 2014;53(7):852–864. doi: 10.3109/0284186X.2014.895036. [DOI] [PubMed] [Google Scholar]
- 24.Wu S., Gan Y., Wang X. PIK3CA mutation is associated with poor survival among patients with metastatic colorectal cancer following anti-EGFR monoclonal antibody therapy: a meta-analysis[J] J. Cancer Res. Clin. Oncol. 2013;139(5):891–900. doi: 10.1007/s00432-013-1400-x. [DOI] [PubMed] [Google Scholar]
- 25.Ulivi P., Capelli L., Valgiusti M. Predictive role of multiple gene alterations in response to cetuximab in metastatic colorectal cancer: a single center study[J] J. Transl. Med. 2012;10:87. doi: 10.1186/1479-5876-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manceau G., Marisa L., Boige V. PIK3CA mutations predict recurrence in localized microsatellite stable colon cancer[J] Cancer Med. 2015;4(3):371–382. doi: 10.1002/cam4.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao X., Lochhead P., Nishihara R. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival[J] N. Engl. J. Med. 2012;367(17):1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo Z.G., Yu Z.Q., Gao X.H. [Association of epithermal growth factor receptor expression and its downstream gene mutation status with radiosensitivity of colorectal carcinoma cell lines in vitro][J] Zhonghua Wei Chang Wai Ke Za Zhi. 2013;16(8):753–758. [PubMed] [Google Scholar]
- 29.Ganesan P., Janku F., Naing A. Target-based therapeutic matching in early-phase clinical trials in patients with advanced colorectal cancer and PIK3CA mutations[J] Mol. Cancer Ther. 2013;12(12):2857–2863. doi: 10.1158/1535-7163.MCT-13-0319-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phipps A.I., Makar K.W., Newcomb P.A. Descriptive profile of PIK3CA-mutated colorectal cancer in postmenopausal women[J] Int. J. Color. Dis. 2013;28(12):1637–1642. doi: 10.1007/s00384-013-1715-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stintzing S., Lenz H.J. A small cog in a big wheel: PIK3CA mutations in colorectal cancer[J] J. Natl. Cancer Inst. 2013;105(23):1775–1776. doi: 10.1093/jnci/djt330. [DOI] [PubMed] [Google Scholar]
- 32.Fu X.H., Chen Z.T., Wang W.H. KRAS G12V mutation is an adverse prognostic factor of Chinese gastric cancer patients[J] J. Cancer. 2019;10(4):821–828. doi: 10.7150/jca.27899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu X., Huang Y., Fan X. Demographic trends and KRAS/BRAF(V600E) mutations in colorectal cancer patients of South China: A single-site report[J] Int. J. Cancer. 2019;144(9):2109–2117. doi: 10.1002/ijc.31973. [DOI] [PubMed] [Google Scholar]
- 34.Souglakos J., Philips J., Wang R. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer[J] Br. J. Cancer. 2009;101(3):465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J.M., Wang Y., Wang Y.L. PIK3CA mutations contribute to acquired cetuximab resistance in patients with metastatic colorectal cancer[J] Clin. Cancer Res. 2017;23(16):4602–4616. doi: 10.1158/1078-0432.CCR-16-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jhawer M., Goel S., Wilson A.J. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab[J] Cancer Res. 2008;68(6):1953–1961. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wee S., Jagani Z., Xiang K.X. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers[J] Cancer Res. 2009;69(10):4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 38.Anderson G.R., Winter P.S., Lin K.H. A landscape of therapeutic cooperativity in KRAS mutant cancers reveals principles for controlling tumor evolution[J] Cell Rep. 2017;20(4):999–1015. doi: 10.1016/j.celrep.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material