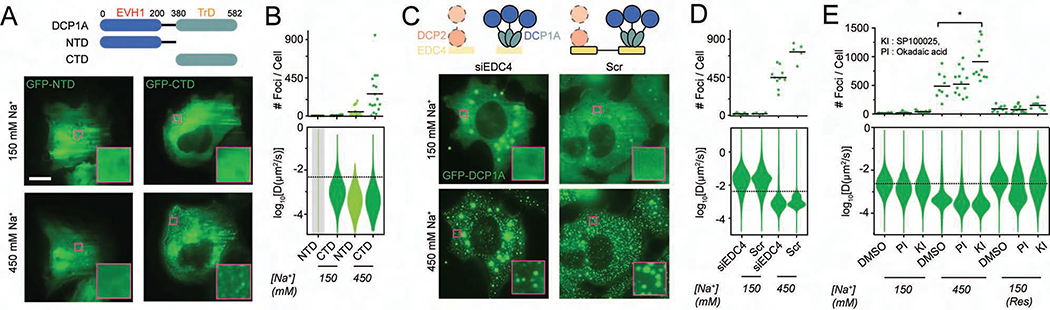
Figure 4. HOPS of DCP1A is dependent on its trimerization domain and modulated by PTMs, but not its interaction with EDC4.
(A) Schematic of full length DCP1A, NTD, or CTD constructs (top, not to scale). EVH1 domain, trimerization domain, and the amino acid numbers are marked. Representative pseudocolored images of U2OS cells (GFP, green) transfected with GFP-NTD or GFP-CTD that were treated with isotonic (150 mM Na+) or hypertonic (300 mM Na+) medium (bottom). Scale bar, 10 μm. (B) Scatter plot of the number of foci per cell (top) and violin plots of diffusion constants associated with DCP1A foci (bottom) imaged in panel A. n = 3, > 5 cells per sample. (C) Schematic of DCP1A, DCP2 and EDC4 in the decapping complex (top, not to scale) in siEDC4 or Scr treatment conditions. Representative pseudocolored images of siEDC4 or Scr siRNA treated UGD cells (GFP, green) treated with isotonic (150 mM Na+) or hypertonic (300 mM Na+) medium (bottom). Scaled as in panel A. (D) Scatter plot of the number of foci per cell (top) and violin plots of DCP1A diffusion constants (bottom), associated with assay represented in C. n = 3, > 5 cells per sample. (E) Scatter plot of the number of foci per cell (top) and violin plots of DCP1A diffusion constants (bottom) within UGD cells that were pre-treated treated with DMSO, KI, or PI, and imaged in isotonic (150 mM Na+) medium, hypertonic (300 mM Na+) medium, or rescued (Res) with isotonic medium after hypertonic treatment. n = 3, > 5 cells per sample. The dotted line in the diffusion plots empirically demarcates high- and low-mobility fractions. See also Figure S4.