Figure 5. Compound IXA4 increases degradation of amyloid precursor protein (APP) mutants.
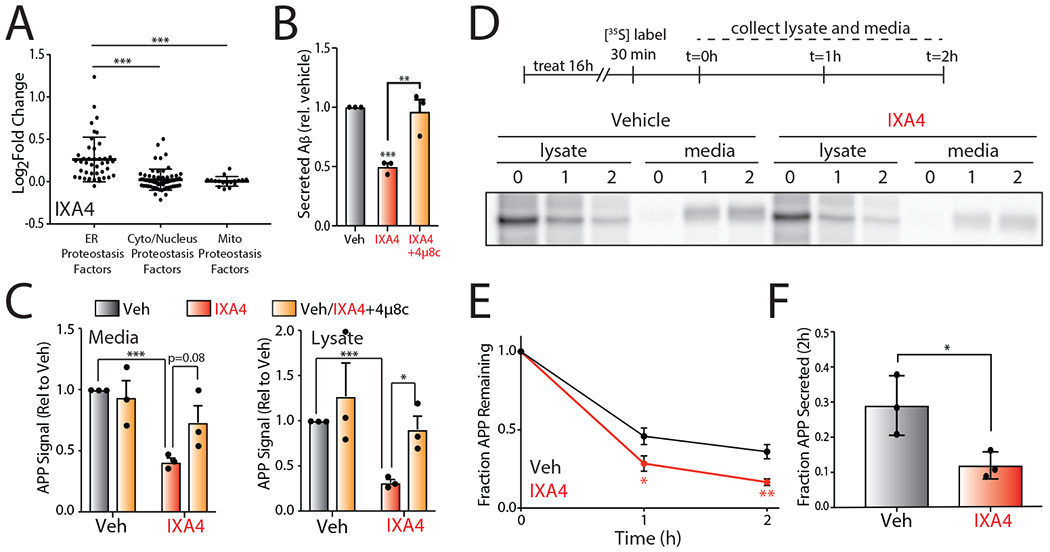
I. Plot of log2 Fold Change mRNA levels from RNAseq in cells treated with IXA4 (10 μM, 4hr) of proteostasis factors found in the ER, cytosol/nucleus, or mitochondria. The composition of these proteostasis genesets is shown in Source Data Table 3.
J. Graph showing relative signal from ELISA of secreted Aβ peptide in conditioned media from CHO7PA2 cells treated with IXA4 (10 μM) in the presence or absence of 4μ8c (32 μM). Cells were pretreated for 18 hrs with compounds. Media was then replaced and conditioned in the presence of compounds for 24 hrs before harvesting conditioned media for ELISA. Secreted Aβ was normalized to that observed in untreated controls. Error bars represent SE for n = 3 replicates. P-values calculated from one-tailed Student’s t-test. **p<0.01, ***p<0.001.
K. Quantification of mutant APP relative to vehicle-treated controls in media and lysate isolated from CHO7PA2 cells treated with IXA4 (10 μM) and/or 4μ8c (32 μM) as in panel B, measured by immunoblotting. A representative immunoblot is shown in Extended Data Fig. 8A. Error bars represent SD for n = 3 replicates. P-values calculated from one-tailed Student’s t-test. *p<0.05, ***p<0.001.
L. Representative autoradiogram showing the [35S] metabolic labeling of mutant APP in CHO7PA2 cells treated with IXA4 (10μM) for 16 hrs prior to 30 min labeling. Media and lysates were collected at 0, 1, or 2 hrs and [35S]-labeled mutant APP was isolated by immunopurification. The experimental protocol is shown above.
M. Plot showing fraction mutant APP remaining at each time point of the metabolic labeling experiment shown in panel D. Fraction remaining was calculated using the following equation: (APP in lysate at time = t + APP in media at time = t) / (APP in lysates at t = 0 + APP in media at t = 0). Error bars represent SD for n = 3 replicates. P-values were calculated from one-tailed Student’s t-test. *p<0.05, **p<0.01.
N. Plot showing fraction of fraction APP secreted at 2 hrs of the metabolic labeling experiment shown in panel D. Fraction secretion was calculated using the following equation: (APP in media at time = t) / (APP in lysates at t = 0 + APP in media at t = 0). Error bars represent SD for n = 3 replicates. P-values were calculated from one-tailed Student’s t-test. *p<0.05.
