Figure 1. Modular assembly of the 50S subunit.
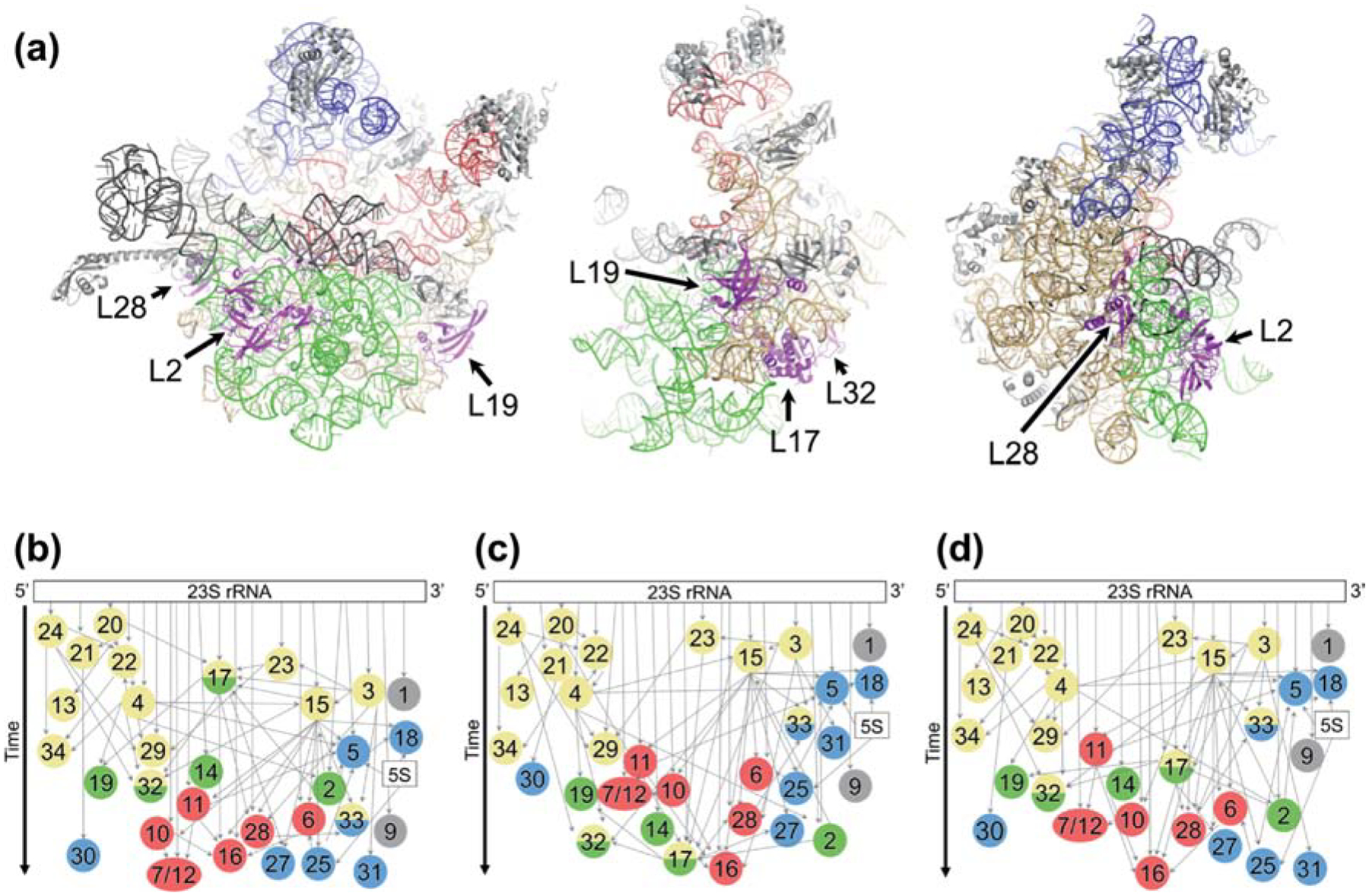
(a) Three views of the structure of the 50S subunit (PDB 4YBB) showing rRNA assembly block 1 (tan), block 2 (blue), block 3 (green), and block 4 (red). Other regions of rRNA, shown in gray, include flexible elements rarely resolved in intermediates. L17 and L17-dependent proteins are highlighted in magenta and labeled. (b-d) LSU assembly maps depicting the relative rates of r protein incorporation by vertical position. Colors in circles represent the block(s) with which each protein is associated, based on experimental evidence (Davis et al. 2016) or inferred from the structure of the mature LSU. Block 1, yellow; block 2, blue; block 3, green; block 4, red; unassigned, gray. Narrow gray arrows, thermodynamic binding dependencies; wide black arrow, time. (b) Map based on data from WT cells grown at 37° C in the absence of amino acid supplementation (Chen and Williamson, 2013). (c) Map based on data from ΔbipA cells grown at 20° C. (d) Map based on data from WT cells grown at 37° C with Lys and Arg supplementation.
