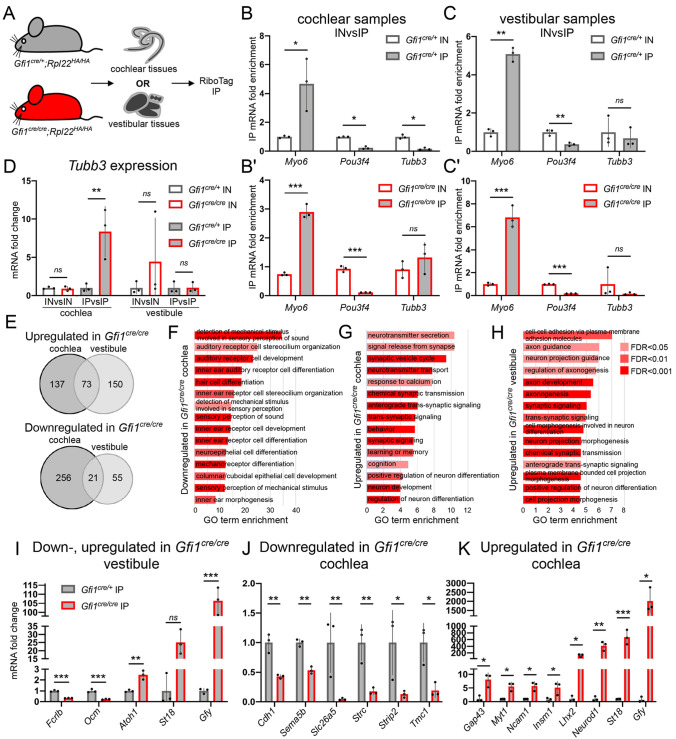
Fig. 2.
Translatome analysis reveals significant upregulation of neuronal mRNA in the HCs of GFI1 mutant mice. (A) P0 cochlear and vestibular tissues from mice expressing Rpl22HA in HCs were collected separately for RiboTag immunoprecipitation. (B,C) In both cochlear (B, Gfi1cre/+; B', Gfi1cre/cre) and vestibular (C, Gfi1cre/+; C', Gfi1cre/cre) samples, immunoprecipitates (IPs) had higher levels of transcripts for the HC-expressed gene Myo6 compared with input (IN), but had lower levels of the transcripts for the mesenchymal-expressed gene Pou3f4. Immunoprecipitates were not enriched with Tubb3 (n=3). Dots represent individual replicates. Data are mean±s.d. (D) Tubb3 is upregulated in the mutant cochlear IPs compared with control IPs (fold change=7.87, P=0.0046), but not in mutant cochlear IN compared with control IN (n=3). (E) Number of genes upregulated and downregulated in the Gfi1cre/cre cochlear and vestibular HCs. (F-H) Top 15 enriched gene ontology (GO) terms from genes downregulated (F) or upregulated (G) in Gfi1cre/cre cochlear HCs, or upregulated in Gfi1cre/cre vestibular HCs (H). (I-K) qPCR validation of dysregulated genes in vestibular (I) and cochlear (J,K) Gfi1cre RiboTag immunoprecipitation samples (n=3). *P<0.05, **P<0.01, ***P<0.001, ns, not significant. Statistical significance assessed by a two-tailed Welch's t-test. Data are mean±s.d.