Abstract
Plasticity of cancer invasion and metastasis depends on the ability of cancer cells to switch between collective and single-cell dissemination, controlled by cadherin-mediated cell–cell junctions. In clinical samples, E-cadherin-expressing and -deficient tumours both invade collectively and metastasize equally, implicating additional mechanisms controlling cell–cell cooperation and individualization. Here, using spatially defined organotypic culture, intravital microscopy of mammary tumours in mice and in silico modelling, we identify cell density regulation by three-dimensional tissue boundaries to physically control collective movement irrespective of the composition and stability of cell–cell junctions. Deregulation of adherens junctions by downregulation of E-cadherin and p120-catenin resulted in a transition from coordinated to uncoordinated collective movement along extracellular boundaries, whereas single-cell escape depended on locally free tissue space. These results indicate that cadherins and extracellular matrix confinement cooperate to determine unjamming transitions and stepwise epithelial fluidization towards, ultimately, cell individualization.
Collective cell migration results from the translocation of multicellular groups, which coordinate front–rear polarity, actomyosin contractility, as well as mechanical and signalling cooperation through cell–cell junctions1,2. In epithelial and endothelial cells migrating as cohesive sheets, cadherin adhesion receptors mechanically connect neighbour cells and, through intracellular adaptors, coordinate actomyosin contractility and multicellular polarity3,4. Moving epithelia during morphogenesis depend on E-cadherin-based cell–cell junctions to form skin, gut, glands and ducts5 and retain active force transmission as well as coordination across multiple cell bodies6. Conversely, in progressing cancers, E-cadherin is variably deregulated as part of the epithelial-to-mesenchymal transition (EMT), which induces molecular reprogramming with destabilized cell–cell junctions7 and increased invasiveness and metastasis8,9. Therefore, E-cadherin is generally considered as a gatekeeper maintaining epithelial differentiation and counteracting oncogenic transformation10,11.
Clinical data indicate that epithelial cancer progression and metastasis can occur with or without E-cadherin deregulation12–14. In invasive ductal carcinomas (IDC) E-cadherin is expressed during collective invasion12 and supports cancer cell survival during metastasis progression15. Collective metastasis invasion zones and circulating tumour-cell clusters retain E-cadherin expression and epithelial differentiation, without undergoing complete EMT11,16–18. However, loss of E-cadherin does not preclude multicellular invasion and metastasis in invasive lobular carcinoma (ILC)12. Thus, the conditions determining how E-cadherin deregulation impacts collective invasion in vivo remain unclear.
As well as by adherens junctions, the ability of moving cells to engage with neighbour cells and move collectively is modulated by microenvironmental conditions. These include cell density19,20, the topology of extracellular matrix (ECM), which guides moving cells21–23, the engagement of alternative, cadherin-independent cell–cell junctions24,25, and effects caused by volume exclusion between cells that align reciprocally while migrating26. Invading mesenchymal melanoma and sarcoma cells, which are typically known to invade individually, can invade collectively when three-dimensional (3D) tissue density is high and the activity of matrix metalloproteinases (MMPs) contribute to tissue alignment20,27. In vivo invading cancer cells preferentially orient along aligned collagen fibres and bundles, vascular structures and nerves23,28–30, and mesenchymal tumour cells move collectively along these confined trails despite their labile cell–cell junctions20,31–33. From a biophysical perspective, in analogy to foams and colloids, there is an analogy between cell density and migration behaviour, termed ‘jamming transition’26,34. Cell jamming occurs at high cell density when cells cannot overcome the yield stress to pass by each other and thus become immobilized34. Accordingly, with decreasing cell density, individual cell kinetics increase and the epithelium fluidizes19,35,36. However, how regulation of adherens junctions interplays with jamming transitions in invasive cancer cells remains unclear.
Here we investigate how cell–cell adhesion and cell confinement affect collective cell motion in vitro, in vivo and in silico. Using molecular-targeted deregulation of E-cadherin expression in breast cancer cells, we tested whether weakening of cell–cell junctions drives invasion plasticity, including a transition from collective to single-cell migration. The data reveal a spectrum of invasion modes in response to cell–cell junction regulation and 3D tissue confinement, thus implicating adherens junction regulation in jamming–unjamming transitions and 3D invasion plasticity.
Results
Collective invasion along interstitial microtracks in human breast carcinoma
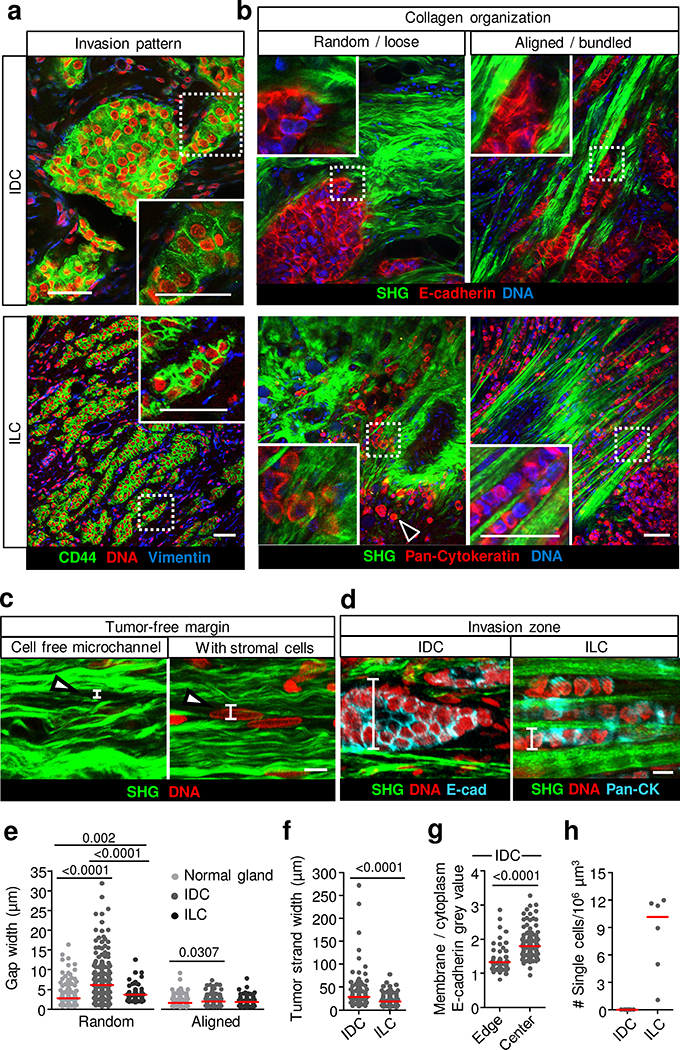
We first addressed the tissue topologies associated with collective invasion in clinical IDC and ILC samples. Invasion zones, identified by expression of cytokeratin and lack of vimentin, consisted of (1) thick multilayered strands in IDC with E-cadherin present along cell–cell junctions; (2) thinner multicellular files that lacked E-cadherin and expressed CD44 along cell–cell contacts; and (3) an additional subset of fully individualized cells in ILC (Fig. 1a,b, Extended Data Fig. 1a and Supplementary Videos 1 and 2), as described12. Multicellular invasion strands resided in cleft-like spaces ranging from 5 to 250 μm in diameter with bordering linear collagen bundles (Fig. 1c–f and Extended Data Fig. 1b,c). Microclefts were also present in tumour-free normal breast tissue and the peritumour stroma, with gap widths ranging from 0.7 to 32 μm (Fig. 1e and Extended Data Fig. 1d,e). Tumour-free and tumour-cell-containing clefts differed in width (Fig. 1e,f), indicating an effect of tumour cells on widening tracks, for example, by mechanical deformation37 and/or proteolytic remodelling22,27. ILC lack E-cadherin expression12, and E-cadherin intensity varied in IDC, with high levels inside strands and lower levels towards the edges (Fig. 1g). Cell individualization was only detected in ILC and absent in IDC (Fig. 1b,h and Extended Data Fig. 1a). Thus, a wide range of E-cadherin expression and cell alignment along three-dimensionally confined tissue clefts are associated with multicellular invasion in IDC and ILC12,38,39. Notably, collective invasion12 and linearized collagen bundles both predict metastatic outcome in breast cancer29,40,41.
Fig. 1. Collective invasion occurs within interstitial tissue tracks.
a, Invasion patterns of CD44- and E-cadherin-positive IDC (top) and CD44-positive, E-cadherin-deficient ILC (bottom). Images are representative of seven and three samples from patients with IDC and ILC, respectively. b, Association of IDC and ILC cell invasion patterns with collagen organization. Examples show randomly organized and aligned collagen fibres detected by second harmonic generation (SHG). Carcinoma cells are detected with E-cadherin (IDC) or pan-cytokeratin (ILC) antibodies. Arrowhead indicates a single carcinoma cell. c, Organization of collagen-rich stroma in a tumour-free zone. Examples show SHG-positive fibrillar collagen and SHG-negative clefts (indicated with arrowheads) without or with stromal cells. d, Organization of collagen-rich stroma in tumour-cell-containing invasion zones. Examples show SHG-positive fibrillar collagen and SHG-negative clefts with carcinoma collective strands. The vertical bars in c,d indicate the width of clefts between collagen bundles. Images in b–d are representative of 7 and 3 samples from patients with IDC and ILC, respectively. e, The distance between SHG-positive collagen fibres with random or aligned organization in tumour-free mammary tissue. Data represent n = 123 random and 97 aligned tissue tracks pooled from 3 independent samples (normal breast tissue), 105 random and 200 aligned tissue tracks from 7 independent samples (tumour-free IDC tissue) and 78 random and 110 aligned tissue tracks from 3 independent samples tumour-free ILC tissue). f, The width of tissue space between collagen bundles filled with carcinoma collective strands in IDC. Data show n = 80 strands pooled from 7 independent IDC samples and n = 87 strands from 3 ILC samples. g, Cell surface E-cadherin level at the edge and centre cells in collective IDC strands. Data represent n = 44 cells (edge) and 110 cells (centre) pooled from 7 patient samples. h, The number of single cells in IDC (7 patient samples) and ILC (3 patient samples) lesions, 1–3 regions per sample. e–h, Scatter plots show the medians (red line) and values representing tissue tracks (e), carcinoma cell strands (f), individual cells (g) or cell counts per region (h). Two-tailed Mann–Whitney test (e–g). Scale bars, 50 μm (a,b); 10 μm (c,d).
Dependence of collective epithelial invasion on tissue density
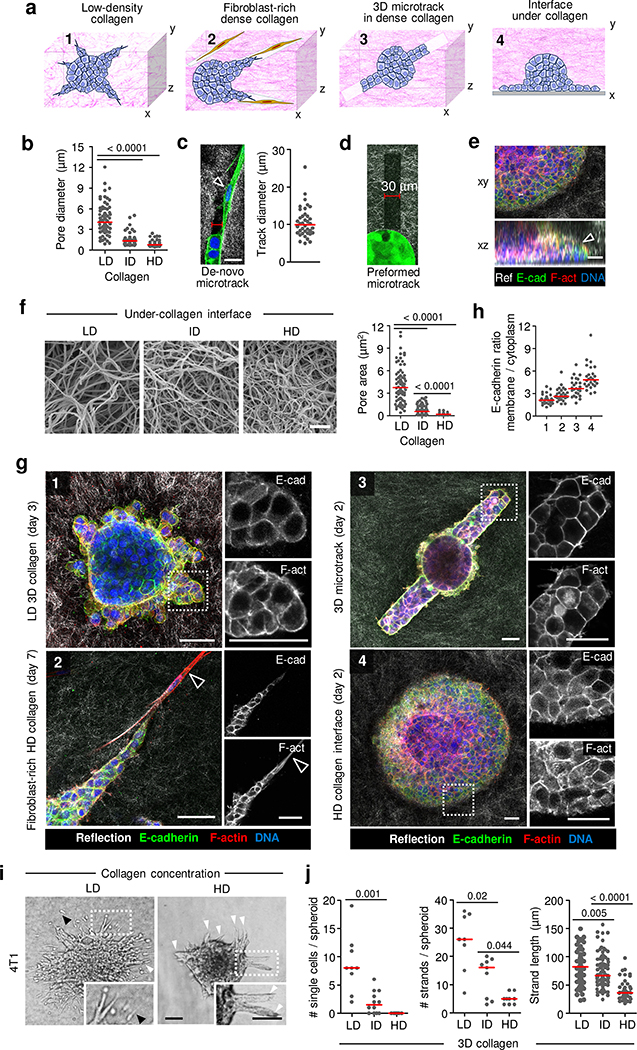
To test how tissue organization affects collective invasion we designed multicellular 3D-spheroid invasion assays with varying collagen topologies and space (Fig. 2a). Reconstituted low-density (0.9–12 μm interfibre space), intermediate-density (0.7–5 μm) and high-density fibrillar collagen (0.46–2.5 μm) (Fig. 2a(i),b, Extended Data Fig. 2f,g and Supplementary Table 1) matched the space range detected in human tumours (Fig. 1e). Fibroblast-conditioned collagen, containing tube-like tracks of variable width22, were used as a cell-derived tissue pattern (Fig. 2a(ii),c), and linear microtracks with defined widths were created by laser ablation37 (Fig. 2a(iii),d). Last, a cleft-like interface model enabled sheet-like migration from spheroids along the interface between culture dish and 3D fibrillar collagen of high or low porosity (Fig. 2a(iv),e,f and Supplementary Table 1). Owing to the lack of lateral confinement, collective interface migration proceeded as continuous sheets rather than finger-like strands.
Fig. 2. Collective invasion of breast carcinoma cells depends on availability of tracks within 3D matrix in vitro.
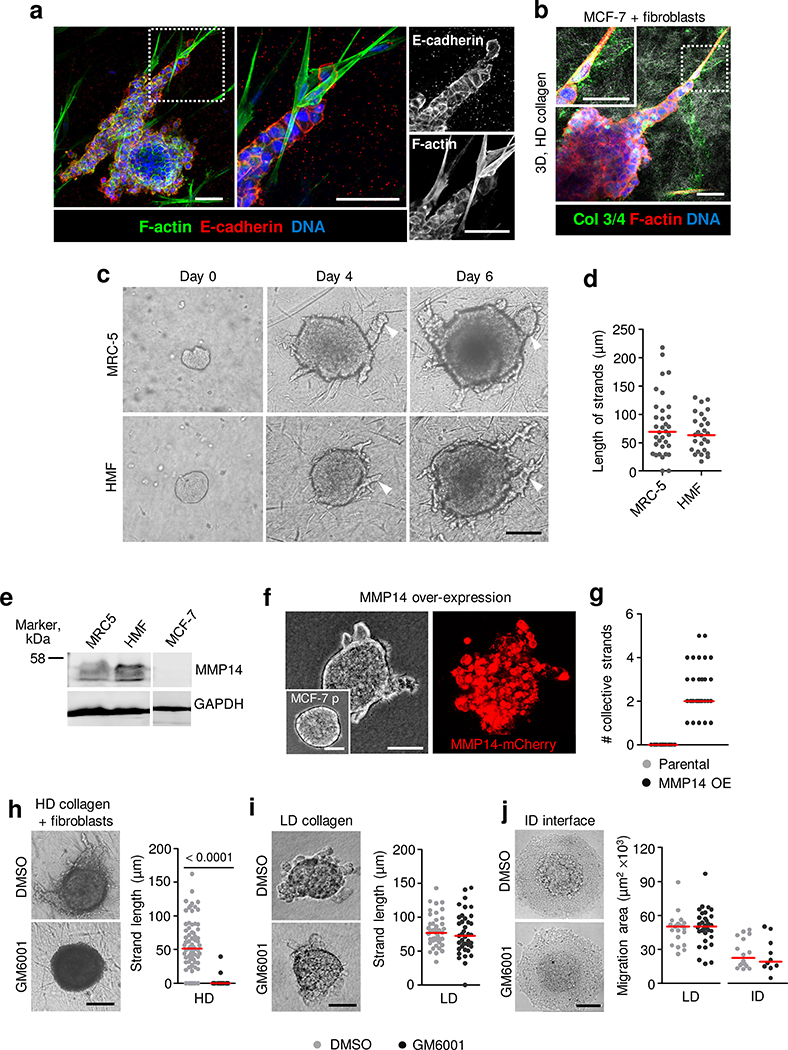
a, Schematic of collagen-based invasion assays used to model MCF-7 cell invasion. b, Pore diameters at different collagen densities, used for 3D invasion of MCF-7 cells. n = 70 (low-density (LD) and intermediate-density (ID)) and 44 (high-density (HD)) pores, pooled from 3 independent experiments. c, Collagen-free de novo fibroblast-generated track at tip of invading MCF-7 cell strand. Arrowhead, fibroblast in microtrack. Quantification of track diameter using the distance between opposing interfaces of the confocal reflectance signal. n = 38 tracks pooled from 3 independent experiments. d, Laser-ablated track in a HD collagen matrix alongside a carcinoma cell spheroid. e, Collective invasion of MCF-7 cells along the interface below a 3D collagen matrix. Arrowhead, leading edge of collective sheet. Images in d,e are representative of 3 independent experiments. Ref, reflectance; E-cad, E-cadherin; F-act, F-actin. f, Scanning electron microscopy of the bottom interface of 3D collagen matrices at different densities (left) and quantified pore diameters (right). n = 72 (LD), 85 (ID) and 71 (HD) pores, 1 experiment. g, Collective invasion of MCF-7 cells cultured in (i) LD collagen, 3 d of culture; (ii)HD collagen cocultured with fibroblasts, 7 d of culture; (iii) HD collagen with tracks, 2 d of culture; (iv) at the interface under HD collagen, 2 d of culture. Arrowheads, fibroblast. Micrographs represent 3 independent experiments. h, E-cadherin membrane/cytoplasm intensity ratio in collectively invading cells after 2–7 d of culture in different matrices. Numbers correspond to conditions described in g. n = 24 (i), 28 (ii), 25 (iii) and 26 (iv) cells per condition, pooled from 3 independent experiments. i, Efficiency of collective invasion in autonomously invasive 4T1 cells in 3D collagen matrices of different density. Black arrowheads, single cells; white arrowheads, collective strands. Representative images from 3 independent experiments. j, Number of single cells or collective strands and length of collective strands from spheroids after 48 h culture in each collagen matrix. n = 9 (LD), 12 (ID) and 9 (HD) (left), and 8 (LD), 9 (ID) and 9 (HD) (middle) spheroids, resulting in n = 93 (LD), 93 (ID) and 45 (HD) (right) collective strands, pooled from 3 independent experiments. Medians (red line) and dots represent individual collagen pores (b,f), collagen microtracks (c), individual cells (h), spheroids (j, left and middle) or collective strands (j, right). Two-tailed Mann–Whitney test. LD, ID and HD collagen matrices are described in Supplementary Table 1. Scale bars, 20 μm (c), 50 μm (e,g), 1 μm (f); 100 μm (i).
We first tested epithelial E-cadherin-positive MCF-7 cells, which lack the expression of membrane type 1-matrix metalloproteinase 1 (MT1-MMP)/MMP14 and the ability to proteolytically remodel ECM (Extended Data Fig. 3e). MCF-7 cells invaded collectively with junctional E-cadherin and F-actin when 3D conditions provided sufficient space, irrespective of the applied 3D model (Fig. 2g(i–iv),h and Extended Data Fig. 2a–d). Collective migration diminished with increasing collagen density in the collagen interface assay (Extended Data Fig. 2e). Cocultured fibroblasts enabled MCF-7 invasion (Extended Data Figs. 2,c,d and 3a–d), and this was inhibited by the broad-spectrum MMP inhibitor GM6001 (Extended Data Fig. 3h). However, at low collagen density, MCF-7 invasion was not inhibited by GM6001 (Extended Data Fig. 3i,j). Thus, MCF-7 cells exploit available ECM space for collective movement. When MT1-MMP/MMP14, the rate-limiting invasion-promoting protease in fibrillar collagen27, was ectopically expressed in MCF-7 cells, collective invasion into high-density collagen was enabled (Extended Data Fig. 3e–g). This indicates that collective invasion of MCF-7 cells depends on either pre-existing or, by means of proteolysis, de novo-generated 3D tissue space.
Single-cell detachment after downregulation of E-cadherin
Besides MCF-7 cells, which retain epithelial differentiation and move only collectively, we further tested highly metastatic 4T1 cells that have undergone mild EMT but retain significant E-cadherin expression and migrate mostly collectively in 3D culture28,42. In spheroid culture, 4T1 cells invaded collectively with E-cadherin present along cell–cell junctions, with occasional single-cell detachment in low- and intermediate-density collagen (Fig. 2i,j). Detached cells retained low levels of E-cadherin and β-catenin (Extended Data Fig. 2h,i), associating E-cadherin regulation with individualization, as reported15.
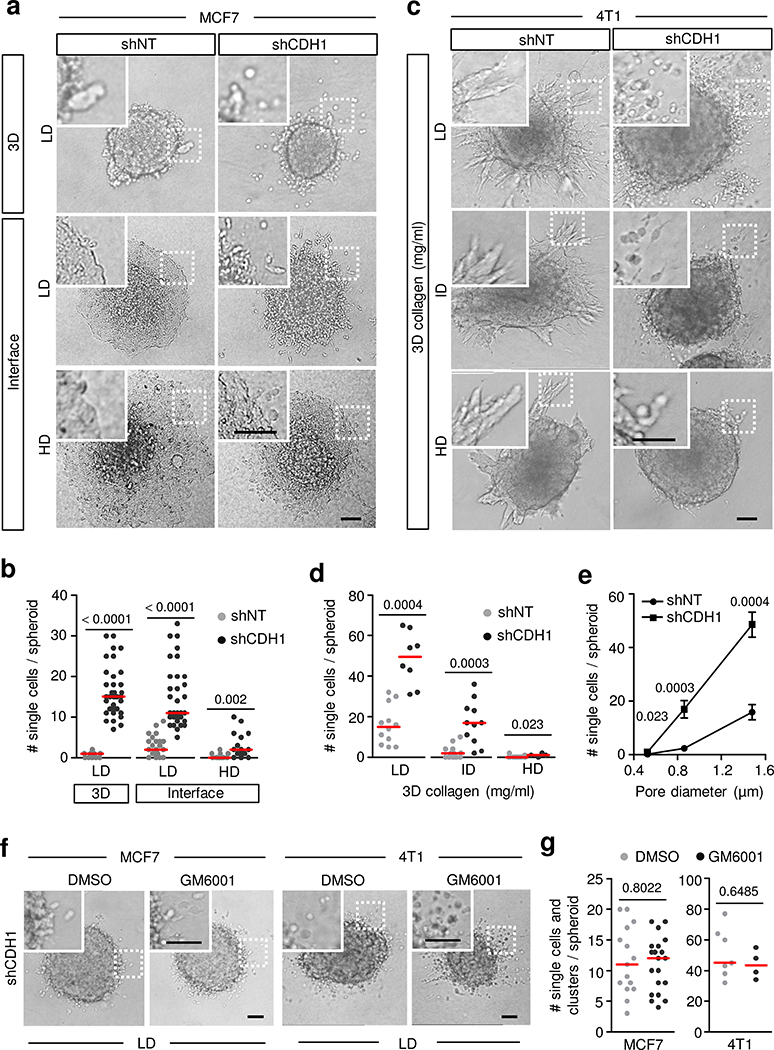
To address how matrix confinement and adherens junction regulation cooperate in controlling collective or single-cell invasion, E-cadherin was stably downregulated in MCF-7 and 4T1 cells, and single-cell transitions were assessed for low- and high-density ECM conditions (Fig. 3a–e and Extended Data Fig. 4a–f). For MCF-7 cells, an established cell model in which E-cadherin level is decreased by more than 90% was used7. For 4T1 cells, expression of the E-cadherin gene (CDH1) was reduced with CDH1 short hairprin RNA (shRNA) (shCDH1) using 5 different shRNA sequences, yielding 4T1-shCDH1 cells with comparable downregulation (85–95%; Extended Data Fig. 4d–f), and shRNA 78 was used subsequently (Supplementary Table 2). Downregulation of E-cadherin in MCF-7 cells caused upregulation of the mesenchymal markers N-cadherin, vimentin and Zeb-2; whereas in 4T1-shCDH1 cells downregulation of E-cadherin decreased levels of α- and β-catenin, but not p120-catenin, without altering mesenchymal marker expression (Extended Data Fig. 4g). Thus, CDH1low MCF-7 and 4T1 cells both retained a partial EMT signature.
Fig. 3. Single-cell detachment after downregulation of E-cadherin requires non-confining matrix conditions in vitro.
a, Invasion modes of MCF-7 cells after knockdown of E-cadherin in LD 3D collagen matrix or in the 3D interface assay overlaid with LD or HD collagen after 48 h culture. Micrographs represent 3 independent experiments. b, The number of single cells released from MCF-7 spheroids in LD 3D collagen matrix and along interfaces formed by LD or HD collagen matrices. Data represent n = 12, 23, 13 (shNT) or 37, 33, 17 (shCDH1) spheroids in 3D LD, interface LD and interface HD conditions, respectively, pooled from 3 independent experiments. c, Efficiency of single-cell dissemination after E-cadherin knockdown in 4T1 cells invading 3D collagen matrices with different densities after 48 h. Example images are from 3 independent experiments. d, The number of single cells released from 4T1 spheroids in 3D matrices of different collagen densities. Data represent n = 12, 14, 15 (shNT) and 8, 11, 4 (shCDH1) spheroids in 3D LD, ID and HD collagen conditions, respectively, pooled from 3 independent experiments. e, Dependency of 4T1 single-cell release of diameter of collagen pores with culture conditions and number of spheroids for quantification as in d. Data are mean ± s.e.m. from n = 3 independent experiments. f, Single-cell dissemination of MCF-7 and 4T1 cells with E-cadherin knockdown in LD 3D collagen matrix after blocking of MMPs with GM6001. Micrographs represent 3 (MCF-7) or 2 (4T1) independent experiments. g, Number of individually invading MCF-7-shCDH1 and 4T1-shCDH1 cells with and without GM6001 in LD 3D collagen matrix after 48 h. Data represent the number of individualized cells from 15 and 20 (shNT) and 7 and 4 (shCDH1) spheroids for DMSO and GM6001, respectively, pooled from 3 (MCF-7) or 2 (4T1) independent experiments. Scatter plots in b,d,g show medians (red lines) and values representing cells per spheroid. Two-tailed Mann–Whitney test. LD, ID and HD collagen matrices are described in Supplementary Table 1. Scale bars, 100 μm (a,c,f).
E-cadherin repression caused single-cell release of MCF-7 and 4T1 cells in low- to intermediate-density collagen, but not in high-density collagen (Fig. 3a–e). Individualization in conditions of low confinement was also induced by the monoclonal antibody SHE78–7, which perturbs homophilic E-cadherin interactions (Extended Data Fig. 5a–f). Combining shRNA-mediated and antibody interference did not further increase single-cell release (Extended Data Fig. 5g,h), indicating that both strategies lowered cell–cell adhesion effectively. Individualization in low-density collagen was not reduced by inhibition of MMP activity with GM6001, suggesting that single-cell dissemination occurred via MMP-independent cell squeezing between collagen fibrils30 (Fig. 3f,g). These results indicate that single-cell detachment is enabled by weakened adherens junctions and sufficient interstitial space.
Collective invasion despite E-cadherin downregulation
Individualization of CDH1low MCF-7 and 4T1 cells was gradually reverted to collective invasion as ECM density was increased (Fig. 3b–e and Extended Data Fig. 5a–c). The number and length of multicellular strands decreased in 4T1-shCDH1 cells when confronted with high-density collagen (Extended Data Fig. 4h,i), but remained unchanged in MCF-7-shCDH1 cells when fibroblasts were present (Extended Data Fig. 4j,k). Thus, the ability to move collectively was regained when the ECM was dense and contained guiding trails. The junctions between CDH1low cells lacked E-cadherin but contained F-actin, with β-catenin retained between MCF-7 but not between 4T1 cells (Fig. 4a–c and Extended Data Fig. 4l). Thus, multicellular invasion persists despite compromised adherens junction organization when tissue confinement is high.
Fig. 4. Variability of cell–cell junction composition and stability in breast cancer cells migrating under low- or high-confinement ECM.
a, β-Catenin (β-cat) localization in MCF-7-shNT or MCF-7-shCDH1 cells in LD or HD interface assay after 48 h (described in Supplementary Table 1). Micrographs represent 3 independent experiments. Dots represent boundary/cytoplasm ratio of E-cadherin and β-catenin (β-cat) intensity of n = 35 (E-cadherin LD), 29 (E-cadherin HD), 27 (β-catenin LD) and 31 (β-catenin HD) for MCF-7-shNT cells, and n = 35 (E-cadherin LD), 30 (E-cadherin HD), 18 (β-catenin LD) and 39 (β-catenin HD) for MCF-7-shCDH1 cells, pooled from 3 independent experiments. b, Adherens junction protein localization in MCF-7 cells invading collectively in HD collagen in coculture with fibroblasts after 7 d. Micrographs represent 3 independent experiments. Dots represent boundary/cytoplasm intensity ratio of n = 26 (E-cadherin), 35 (β-catenin) and 36 (p120-catenin (p120)) for MCF-7-shNT cells, and n = 30 (E-cadherin), 35 (β-catenin) and 36 (p120-catenin) for MCF-7-shCDH1 cells, pooled from 3 independent experiments. c, β-catenin localization in collectively invading 4T1-shNT or 4T1-shCDH1 cells after 48 h. Micrographs represent 3 independent experiments. Dots represent the boundary/cytoplasm intensity ratio from n = 22 (E-cadherin LD), 21 (E-cadherin ID), 35 (β-catenin LD) and 41 (β-catenin ID) for 4T1-shNT, and n = 20 (E-cadherin LD), 20 (E-cadherin ID), 35 (β-catenin LD) and 41 (β-catenin ID) for 4T1-shCDH1 cells, pooled from 3 independent experiments. d,e, Combined downregulation of indicated adherens junction proteins using short interfering RNA (siRNA) in MCF-7-shCDH1 cells in LD or HD interface assay (48 h culture). siNT, non-targeting siRNA; siβ-cat, β-catenin siRNA; siP120, p120-catenin siRNA. d, Bright-field micrographs representative of 3 independent experiments. White arrowheads, detached cells; black arrowheads, collectively moving cells. e, Number of single cells and detached clusters in LD interface conditions. N-cad, N-cadherin; P-cad, P-cadherin; α-cat, α-catenin. Dots represent n = 40 spheroids for cells treated with non-targeting siRNA and siRNA targeting E-cadherin, N-cadherin, P-cadherin, α-catenin and β-catenin, and n = 44 spheroids for siP120, pooled from 3 independent experiments. f,g, Effect of ECM density on migration pattern in MCF-7-shCDH1 cells treated with siP120 in a LD interface assay. f, β-Catenin localization in collectively and individually invading MCF-7-shCDH1 cells in the interface assay. Arrowheads, cell–cell junctions. Micrographs are representative of 3 independent experiments. g, β-Catenin boundary/cytoplasm intensity ratio in individually or collectively migrating cells in LD or HD collagen interface conditions. Dots show intensity ratio from n = 35 cells per condition pooled from 3 independent experiments. Scatter plots show the medians (red lines) and values from cells (a–c,g) or spheroids (e). Two-tailed Mann–Whitney test. Scale bars, 50 μm (a–c,f) and 100 μm (d).
To further investigate the prediction that ECM confinement dictates collective migration of cells with negligible or fully absent cell–cell adhesion, we transiently downregulated adherens junction proteins in MCF-7-shCDH1 cells, including E-cadherin, N-cadherin, P-cadherin, α-catenin, β-catenin and p120-catenin, and measured the migration mode in the collagen interface model (Fig. 2a(iv)) with high or low ECM confinement. Co-repression of E-cadherin and β-catenin or p120-catenin indeed enhanced individualization in low-density collagen, but collective sheets migrated when collagen density was high (Fig. 4d,e). Since p120-catenin is non-redundant in stabilizing adherens junctions43 and its downregulation was most effective in releasing single cells, we tested whether high confinement rescued cell–cell junctions. β-catenin was diminished in individually moving cells after detachment, but was retained between cells moving as sheet along high-density collagen (Fig. 4f,g).
In silico analysis of invasion plasticity under confinement
To identify the minimal requirements of cell–cell adhesion and ECM confinement in controlling collective or single-cell migration, we applied a lattice-gas cellular automaton in silico model44,45, in which cells are represented as point particles moving between positions on a 2D lattice (Fig. 5a). We simulated a multicellular sheet moving under a deformable ECM network (Fig. 5b) mimicking the interface migration assay (Fig. 2a(iv)), in which leader cells open the space between the collagen-coated glass bottom and the overlaid 3D collagen meshwork (Fig. 2e). In the model, cells change their velocity according to a stochastic interaction rule that incorporates cell–cell adhesion and steric hindrance by ECM fibres, enabling study of the minimum requirements governing transitions between individual and collective migration46,47.
Fig. 5. In silico modelling of collective and single-cell dynamics in dependence of cell–cell adhesion and extracellular confinement.
a, On a hexagonal lattice (1) cells assume discrete velocities c0−8 on the lattice nodes (2), where c0−5 correspond to movement to the next-neighbour nodes and c6−8 = 0 correspond to no movement (‘resting state’). (3) Dynamics: during a discrete time step t → t + 1, cells stochastically change their velocity according to the interaction rule (R), followed by a deterministic propagation (P) of cells with non-zero velocity to the respective next-neighbour node. (4) Cell–cell interactions: a single cell (orange) interacts with cells on a next-neighbour node (black). Cell–cell adhesion couples cells mechanically resulting in the alignment of cell velocities and preventing cell detachment. (5) Extracellular confinement depends on the local ECM density (grey, high; white, low) and reduces cell speed. b, Gap widening by the moving front of the sheet. Principle (cartoon) and normalized cell- and ECM densities projected along the invasive front for cells with strong cell–cell adhesion moving towards high ECM density. c, Average cell speed at the invasion front in response to confinement. Arbitrary units (a.u.). Data in b,c are mean ± s.e.m. at the final time point from 5 independent simulations. d, Topology of migration patterns after 200 time steps for low and high cell–cell-adhesion strength and ECM density. Vectors correspond to local cell flux. Colour indicates velocity correlation between nearest neighbours, with anti-correlated (red) or correlated (green) movement. Blue circles, single cells. e, Cell individualization (colour) and next-neighbour velocity correlation (z-axis) as a function of extracellular substrate density (x-axis, log scale) and cell–cell adhesion strength (y-axis, log scale). Data are means from 5 independent simulations. Regions denote principal response types: (1) single-cell release with minimal next-neighbour velocity correlations; (2) lack of single-cell release and high velocity correlation; (3) lack of single-cell release and minimal velocity correlation.
Decreasing cell–cell adhesion and extracellular confinement accelerated migration (Fig. 5c). Likewise, the velocity correlation between cell neighbours, that is the alignment of velocity vectors, and the probability of individualization were dependent on cell–cell adhesion and extracellular confinement (Fig. 5d and Supplementary Video 3). The probability of cell detachment from the sheet was highest at low ECM density and low cell–cell adhesion, and movements of individual cells were uncorrelated (Fig. 5e, region 1). Strong cell–cell adhesion limited individualization, but supported collective movement with strong velocity correlations between neighbour cells (Fig. 5d,e, region 2). When extracellular confinement was high, even negligible cell–cell adhesion strength failed to support single-cell release, and cells moved as a sheet (Fig. 5d); however, nearest-neighbour velocity correlations remained minimal compared with collective movement with strong cell–cell adhesion (Fig. 5e, region 3). Thus, with decreasing cell–cell adhesion, strongly correlated movement undergoes a dual-step transition to individualization: (1) fluid-like collective sheets with uncorrelated movement when cells move along high extracellular confinement, and (2) individualization and gas-like dissipation when extracellular confinement is low.
Epithelial fluidization and individualization after labilizing adherens junctions
We aimed to confirm that adherens junctions and extracellular confinement jointly define the stepwise fluidization of collective migration in 3D ECM culture and tracked CDH1high and CDH1low 4T1 cells emigrating from spheroids maintained in the interface assay (Fig. 2a(iv)). We tested whether increasing ECM confinement restores collective motion and cell–cell cooperation.
In low-density collagen (2 mg ml−1), migrating CDH1low cells individualized towards the outward edge (Extended Data Fig. 6a); however, in high-density collagen (6 mg ml−1) these cells moved as multicellular sheet (Fig. 6a and Extended Data Fig. 6b,c). CDH1low cells accelerated after E-cadherin downregulation, and the highest speed was reached when collagen density was low (Fig. 6b and Supplementary Videos 4 and 5). Despite high ECM confinement, 4T1-shCDH1 cells elongated and developed gaps between cells, whereas 4T1-shNT cells were less elongated and moved as gap-free, cohesive sheets (Fig. 6a (white gap areas), c,d). Sheet-like streams of 4T1-shNT cells produced linear paths (Fig. 6e, left and Supplementary Video 6) with high spatial and velocity correlation (Fig. 6f and Extended Data Fig. 6d–f). This correlation reflects the similarity of speed and direction between neighbouring cells. By contrast, the paths of 4T1-shCDH1 cells were disjointed, and spatial and velocity correlation were strongly diminished (Fig. 6e,f and Extended Data Fig. 6d–f). The spatial and velocity correlation of 4T1-shCDH1 cells migrating along the high-density collagen interface, as a signature of randomness, were statistically identical to the correlations between CDH1low cells, which were fully individualized at the invasion front in low-density collagen (Fig. 6g,h). This suggested that although CDH1low cells moved as near-confluent sheet with F-actin along cell–cell connections (Fig. 6g and Extended Data Fig. 6b,c), there was almost no path coordination, similar to fully individualized cells. Thus, confinement did not restore next-neighbour coordination.
Fig. 6. Types of collective dynamics in dependence of E-cadherin status and ECM confinement.
a. Morphology of 4T1-shNT or 4T1-shCDH1 cells moving along a collagen interface (6 mg ml−1) after 48 h of culture. White lines, cell boundaries; white areas, gaps between cells identified by image segmentation. b, Velocity at the outward edge. n = 15,827 (low-density collagen) and 10,273 (high-density collagen) position changes within 1 h for shNT; n = 17,496 (low-density collagen) and 11,526 (high-density collagen) position changes within 1 h for shCDH1; data pooled from 10 spheroids per condition (2 independent experiments). In box plots, the black line shows the median, box edges represent 25th and 75th percentiles, and whiskers extend to maximum and minimum values. P < 10−6 for all possible comparisons; two-sided Kolmogorov–Smirnov test. c,d, Cell elongation during migration. Dots, mean aspect ratio per spheroid (2 data points per spheroid, 1 representing the migration front and 1 the mid zone) pooled from 3 spheroids per condition from 1 experiment. Black line, correlation of aspect ratio and standard deviation of aspect ratio based on geometrical constraints51. P = 2.5 × 10−13, two-sided Kolmogorov–Smirnov test. d, Aspect ratios at the invasion edge (5–6 outer cell rows) below a 3D collagen matrix (6 mg ml−1). Dots represent n = 2,318 (shNT) and 1,230 (shCDH1) cells pooled from 3 spheroids per condition from 1 experiment. In box plots, lines show medians, small squares represent mean values, box edges represent 25th and 75th percentiles, and whiskers extend to maximum and minimum values. P = 8.56 × 10−11; two-sided Kolmogorov–Smirnov test. e, Migration tracks (blue lines, 6 h period). f, Modes of collective migration, expressed as spatial correlation of the velocity direction (time interval: 27–54 h). Lines represent means from n = 10 spheroids per condition (2 independent experiments). Fisher z-transformation for each x point; difference between edge versus mid zone for the same collagen density is not significant. g,h, Morphology (g) and spatial correlation of the velocity direction (h) at the invasion edge (48 h end point). Data in h are pooled means from n = 10 spheroids per condition (2 independent experiments). i, Zone-related PIV (left) and representative velocity and vorticity map of H2B–mCherry positive cell nuclei (right). Ω, vorticity, defined as the curl (∇) of the velocity (v); R, distance from spheroid center. j,k, Local vorticity and velocity profiles for shNT and shCDH1 cells (43–48 h period). j, Representative velocity and vorticity maps. In k, lines represent means from n = 10 spheroids per condition (2 independent experiments). Micrographs in (a,e,g,i) are representative of n = 10 spheroids (2 independent experiments). Detailed statistical analysis of these data is described in Extended Data Fig. 6g. Low (LD), intermediate (ID) and high (HD) density collagen matrices are described in Supplementary Table 1. Scale bars, 100 μm.
To confirm this unexpectedly strong fluidization or unjamming of cells within the sheet by an orthogonal approach, we detected motion by subcellular-feature tracking using zonally defined particle-imaging velocimetry (PIV). As readouts, we compared the velocity of cells and their vorticity distribution in the spheroid core, mid zone of the sheet and the invasion front (Fig. 6i,j). Vorticity is defined as the curl of the velocity field and describes the local rotation of objects. In a 2D fluid, the vorticity is equal to twice the local angular velocity at each point. In CDH1high cells, velocity increased towards the outward front of the moving sheet, and further increased when CDH-1 was downregulated (Fig. 6k). Vorticity was low in 4T1-shNT cells moving along high confinement (Fig. 6j,k and Supplementary Video 7), confirming well-correlated directed streaming motion in the multicellular sheet. Lowering confinement increased the vorticity of 4T1-shNT cells moderately, but downregulating E-cadherin caused uncorrelated vector fields independent of collagen density (Fig. 6j,k, Extended Data Fig. 6g and Supplementary Video 7). Nucleus-based cell tracking and PIV thus both reveal a loss of next-neighbour correlation in moving multicellular sheets in CDH1low cells.
Plasticity of invasion after deregulating adherens junctions in vivo
To clarify whether the transition from collective movement to single-cell movement after E-cadherin deregulation also depends on ECM space constraints in vivo, we implanted 4T1 cells expressing histone 2B (H2B)–mCherry and Lifeact–enhanced green fluorescent protein (eGFP) as multicellular spheroids into the mammary fat pad and subsequently imaged and tracked them using multiphoton microscopy (Fig. 7a, left). Control 4T1 cells invaded collectively, with E-cadherin present along cell–cell junctions (Extended Data Fig. 7a). Multicellular strands tended to invade the collagen-rich stroma (Fig. 7b, arrowheads) and were surrounded by vimentin-positive stromal cells (Extended Data Fig. 7a), as described28 and in accordance with collective invasion detected in patient samples12 (Fig. 1). Collective invasion in 4T1-shCDH1 tumours was associated with collagen-rich densely organized stroma (Fig. 7b,c; median pore area 133 μm2), similar to control tumours, and was accompanied by enhanced individualization (Fig. 7d). In control tumours, single-cell release was restricted to niches with larger pores (area 423 μm2), whereas 4T1-shCDH1 cells individualized in regions of lower porosity (area 162 μm2) (Fig. 7b,c). Thus, E-cadherin restrained individualization, and reducing E-cadherin expression favoured single-cell dissemination via pores accommodating the cross-section of moderately deformed cells30 (100–150 μm2).
Fig. 7. Invasion mode depends on ECM confinement in vivo.
a, Mammary imaging model to monitor carcinoma tissue invasion and analysis of spontaneous metastasis. CK-8, cytokeratin-8; IF, immunofluorescence; IHC, immunohistochemistry. b, Collective invasion and single-cell dissemination of control (shNT) or E-cadherin knockdown (shCDH1) 4T1 cells expressing H2B–mCherry and Lifeact–eGFP in tissue subregions. Arrowheads, collective strands. Images are representative of 5 mice per condition. c, Number of detached single cells and associated matrix density in 4T1-shNT or 4T1-shCDH1 lesions. The red line represents the median and dots show average areas from 2–10 collagen pores (SHG signal), each surrounding n = 25 strands and 12 single cells (shNT) and 15 strands and 14 single cells (shCDH1) pooled from 6 spheroids from 4 mice (shNT grouped, shNT single), 7 spheroids from 4 mice (shCDH1 grouped) and 7 spheroids from 7 mice (shCDH1 single). Two-tailed Mann–Whitney test. d, Number of single cells per spheroid for the conditions described in c. Dots show the average number of single cells per spheroid from n = 17 (day 4) and 5 (day 8) spheroids (shNT) or 16 (day 4) and 8 (day 8) spheroids (shCDH1), pooled from 4 mice per condition. Two-tailed Mann–Whitney test. e, Images from time-lapse microscopy of 4T1 cell nuclei in the invasion zone of shNT and shCDH1 tumours and overlaid yellow tracks for a 120 min observation period (maximum intensity projection, 100 μm z-depth). Micrographs represent 3 mice per condition. f, Spatial correlation of the velocity direction obtained by single-nucleus tracking. Lines show the average of 3 spheroids per condition from 3 independent mice. Two-sided Student’s t-test using Fisher z-transformation. g, Mean squared displacement of cells in independent invasion zones (solid lines) from 3 independent mice per condition and guidance lines for power laws with similar exponents (dashed lines). Scale bars, 50 μm.
The tracks of collectively moving 4T1-shNT cells in vivo were linear and largely in parallel, analogous to an active nematic phase48 with partial fluidization in one direction (Fig. 7e, left (yellow trajectories) and Supplementary Video 8). Accordingly, the velocity between adjacent neighbours was correlated (Fig. 7f), resulting in directionally persistent migration as a stream (Fig. 7g). Conversely, 4T1-shCDH1 cells in multicellular regions, before individualization, were mobile (Fig. 7e, right (yellow trajectories)) but did not exhibit next-neighbour velocity correlation (Fig. 7f) and, accordingly, developed decreased migration persistence (Fig. 7g). Thus, the trajectory fields of CDH1high cells in vitro and in vivo were viscous fluid-like, as in collectively moving epithelial monolayers and 3D embryonic tissue49,50, and this correlated motion was strongly perturbed by E-cadherin downregulation.
Lack of increased metastasizing ability after deregulating adherens junctions
To address whether E-cadherin deregulation supports distant metastasis, as predicted from EMT models8,9, or derails metastasis, similar to E-cadherin-deficient variants in vivo15, we orthotopically implanted control MCF-7 cells expressing non-targeting shRNA (MCF-7-shNT) or MCF-7-shCDH1 cells and 4T1-shNT or 4T1-shCDH1 cells and quantified the incidence of spontaneous micro- and macro-metastasis in the lungs using microscopy (Fig. 7a). Metastases were detected as E-cadherin-positive (MCF-7-shNT) and vimentin- (MCF-7-shCDH1) or cytokeratin-positive cells (4T1) (Extended Data Fig. 7b). Reducing E-cadherin expression in MCF-7 cells accelerated primary growth (Extended Data Fig. 7d) but did not induce lung metastasis (Extended Data Fig. 7e). In 4T1-shCDH1 tumours, both primary growth and lung metastasis were reduced by between two and three fold compared with control tumours (Extended Data Fig. 7d,e; marginally significant). Thus, lowering of E-cadherin expression in low or highly metastatic mammary tumours, although facilitating single-cell release, did not enhance distant metastasis.
Discussion
In this study, we identify at least two modes of epithelial collective cancer invasion with distinct stability and composition of cell–cell adhesions, dependence on environmental confinement and propensity to release single cells. Multicellular dynamics depend either on cell-intrinsic cadherin-based cohesion2 or mechanical confinement, which delimits the tissue space, imposes high local cell density and thereby forces multicellular movement between tissue interfaces19,20,31,35,51. Irrespective of tissue confinement, mature adherens junctions maintain high cell density; this enables cancer cells to stream as an active nematic fluid48 (Extended Data Fig. 7f), the movement of which is highly directed and correlated, similar to other fluidized epithelial and endothelial tissues35,36,52. Mature adherens junctions further limit single-cell detachment, similar to non-transformed epithelial and endothelial cells moving cohesively in unconfined environments52,53.
After lowering E-cadherin expression under conditions of high cell density caused by tissue confinement, multicellular migration occurs despite the molecular inability to hold cells together by stable adherens junctions. Here, collective migration encompasses large variations in EMT status, composition of cell–cell contacts and next-neighbour coordination. As an extreme case, CDH1low 4T1 cells move as multicellular sheets along an interface of dense collagen, but their path kinetics resemble those of fully individualized, unrestricted, randomly moving cells54,55 (Extended Data Fig. 7f, active fluid). Thus, wetting-induced CDH1low cell streams behave similar to a fluid and exhibit unjammed, random motion56. CDH1low cells further develop more elongated shapes and ultimately decondense as single cells with gas-like behaviour when 3D ECM topology offers sufficiently large space (Extended Data Fig. 7f, gas-like). Here, gaps and pore diameters exceeding 5–15 μm readily accommodate the single-cell cross-section after deformation20,30. These results indicate that weakening of cell–cell adhesion supports cell elongation, unjamming and epithelial fluidization.
These results are in contrast with previous in silico analyses on the unjamming of dense cell clusters, with volume fraction close to one and without position change (slippage) between neighbour cells34. In this model, cell tension and cell–cell adhesion determine the cell shape, leading to the prediction that cell elongation and unjamming result from increased cell–cell interaction strength34. In 3D tissues and the in silico simulation used in our work, cell–matrix contacts enable cell slippage, and this may support epithelial fluidization after lowering E-cadherin and cell–cell junction stability, thus reducing cell–cell coordination and inducing an unjamming transition. Consequently, with cell–matrix interactions intact, lowering cell–cell adhesion increased cell speed, reduced coordination between adjacent cells, intercellular gap formation and, eventually, single-cell detachment. This suggests that jamming–unjamming transitions are jointly controlled by cell–cell and cell–matrix interactions and cell slippage.
Although single-cell detachment was enhanced after E-cadherin downregulation in porous matrix in vitro, in vivo and in silico, this did not facilitate spontaneous distant metastasis in vivo. This result is consistent with findings that reduced expression or loss of E-cadherin does not necessarily result in distant metastasis15,17,57. Thus, a constitutive CDH1low state favours local cell individualization and partial EMT, but on its own is not sufficient to enhance distant metastasis.
Beyond cancer invasion, ECM confinement may mechanically regulate collective and single-cell invasion in other contexts. In normal mammary epithelium cultured in 3D-reconstituted basement membrane, collective dissemination persists after E-cadherin deletion and perturbation of junctional organization58. Loss of E-cadherin in a mouse model of ILC initiates tumours, but tissue invasion occurs by multicellular strands11 and similar multicellular strands and clusters invade along 3D tissue interfaces in human ILC12 or engineered mouse models of pancreatic and other cancers undergoing partial EMT59. Tissue confinement further supports multicellular motion in melanoma models, and similar mechanisms may control cooperation between cranial neural crest cells moving in dense 3D tissue20,31,33,60. In aggregate, these results support a broadened concept of jamming–unjamming transitions in which adherens junction stability and ECM confinement cooperate to determine (1) jammed and immobile, solid-like behaviour; (2) correlated partially fluid streams of active nematics; (3) random, uncorrelated fluid behaviour; and, ultimately, (4) individualized gas-like dissemination (Extended Data Fig. 7f). Understanding how tissue fluidizes in a stepwise manner will be useful for conceptualizing different types of invasion plasticity and transitions from collective to single-cell migration, including gastrulation, wound healing, cancer invasion and metastatic escape.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41556-020-0552-6. Received: 28 July 2018; Accepted: 30 June 2020.
Methods
Antibodies and reagents
The following antibodies were used: anti-human E-cadherin (3195, lot 13; rabbit monoclonal clone 24E10, Cell Signaling Technologies; 1:300); anti-human E-cadherin (13–5700 lot 73949505A; mouse monoclonal, clone SHE78–7, ThermoFisher; 1:100); anti-human E-cadherin (610182, lot 80098; mouse monoclonal clone 36/E, BD Biosciences; 1:100); anti-mouse E-cadherin (U3254, lot 044M4786; rat monoclonal clone DECMA-1, Sigma-Aldrich; 1:250); anti-mouse N-cadherin (610920, lot 3331780; mouse monoclonal clone 32/N-cadherin, BD Biosciences; 1:100); anti-human or mouse α-catenin (C2081, lot 0000085292; rabbit polyclonal, Sigma-Aldrich; 1:1,000); anti-human MMP14 (MAB 3328, lot LV1580848; mouse monoclonal clone LEM-2/15.8, Merck Millipore; 1:500); anti-human β-catenin (610154, lot 2300995; mouse monoclonal clone 14/beta-catenin, BD Biosciences; 1:200); anti-human β-catenin (ab6302, lot GR139815–3; rabbit polyclonal, Abcam; 1:1,000); anti-mouse p120-catenin (610134, lot 70220; mouse monoclonal clone 98/pp120, BD Biosciences; 1:200); anti-human cytokeratin-8 (ab53280, lot GR-3241174–4; rabbit monoclonal clone EP1628Y, Abcam; 1:2,000); anti-bovine wide-spectrum cytokeratin (ab9377, lot GR3174686–1; rabbit polyclonal, Abcam; 1:100); anti-human vimentin (ab24525, lot GR457716; chicken polyclonal, Abcam; 1:300); anti-human vimentin (MA5–14564, lot QF2041991A; rabbit monoclonal clone SP20, human specific, ThermoFisher; 1:300); anti-human CD44 (HPA005785, lot number unknown; rabbit polyclonal, Sigma-Aldrich; 1:300); anti-human collagen IV (PA1–28534, lot number unknown; rabbit polyclonal, ThermoFisher; 1:200); anti-human Zeb-2 (ABT332, lot VP1403283; rabbit polyclonal, Merck Millipore; 1:1,000); anti-human β-tubulin (AB 2315513, lot number unknown; mouse monoclonal clone E7, Developmental Studies Hybridoma Bank, University of Iowa; 1:10,000); anti-human GAPDH (2118, lot number unknown; rabbit monoclonal clone 14C10, Cell Signaling Technologies; 1:10,000). The lot numbers of four antibodies (anti-human CD44, anti-human collagen IV, anti-human β-tubulin and anti-human GAPDH) were not documented in the in-house database and remain unknown. Secondary antibodies were cross-adsorbed goat anti-mouse, anti-rat, anti-chicken, anti-rabbit IgG (H+L) conjugated with Alexa Fluor 488, 546 or 647, or donkey anti-rat IgG conjugated with Alexa Fluor 488 (ThermoFisher; 1:200). F-actin was visualized with Alexa Fluor 568-conjugated phalloidin (ThermoFisher; 1:200). 4′,6-diamidino-2-phenylindole (DAPI) (Roche diagnostics; 1 μg ml−1) was used for nuclear labelling.
Cell lines and culture
Human mammary fibroblasts (HMF) (ScienCell Research Laboratories, cat no. 7630) and human lung fibroblasts MRC-5 (CCL-171, ATCC) were maintained in DMEM supplemented with 10% fetal bovine serum (Sigma-Aldrich), penicillin (100 U ml−1, PAA), streptomycin (100 μg ml−1, PAA), 1 mM sodium pyruvate and 1 mM l-glutamine (both from ThermoFisher). MCF-7 (HTB-22, ATTC) cells were cultured in the same medium additionally supplemented with insulin from bovine pancreas (10 μg ml−1, Sigma-Aldrich). 4T1 cells (CRL-2539, ATCC) were grown in RPMI media (ThermoFisher) supplemented with 10% fetal bovine serum (Sigma-Aldrich), penicillin (100 U ml−1, PAA) and streptomycin (100 μg ml−1, PAA), 1 mM sodium pyruvate and 1 mM l-glutamine (both from ThermoFisher Scientific). The identity of 4T1 and MCF-7 cells was verified by short tandem repeat DNA profiling (IDEXX BioResearch). No mammalian interspecies contamination was detected. Cells were routinely tested for mycoplasma contamination (MycoAlert, Lonza). All cell lines were negative for mycoplasma contamination. To generate MCF-7 cells expressing eGFP or H2B–eGFP, lentiviral particles containing pLenti6.2v5-eGFP or pLenti6.2v5-H2B-eGFP (Clontech), respectively, were used, followed by blasticidin selection (5 μg ml−1) and cell sorting for GFP-expressing cells (FACS Aria SORP, Becton Dickinson). Lentiviral particles were produced in HEK-293FT human embryonal kidney cells (ThermoFisher; Cat: R70007) using the ViraPower expression system (Life technologies). To generate 4T1 cells expressing H2B–mCherry and LifeAct–GFP, lentiviral particles containing pLenti6.2v5-H2B-mCherry (a gift from A. J. C. de Groof, Nijmegen, The Netherlands) and pLentiCMV-MCS-LifeAct-GFP vector (a gift from O. Destaing, Institut Albert Bonniot, Grenoble, France) were used. Stable clones were selected with blasticidin (5 μg ml−1) and puromycin (3 μg ml−1) followed by cell sorting and gating on the population expressing both H2B–mCherry and Lifeact–eGFP. For MMP14 overexpression, MCF-7 cells were transiently transfected with a MT1-MMP–mCherry construct61 (a gift from S. Linder, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany) and Lipofectamine 2000 (ThermoFisher). All cell lines were maintained at 37 °C in a humidified atmosphere supplied with 5% CO2.
Transient and stable downregulation of gene expression
For transient siRNA-mediated knockdown of adherens junction proteins, cells were exposed for 24 h to four independent pooled on-target plus siRNAs or non-targeting control siRNA (SMARTpool; ThermoScientific Dharmacon; 25 or 50 nM) (Supplementary Table 2) in Opti-MEM serum-reduced medium using Lipofectamine 2000 reagent (both from ThermoFisher), followed by incubation in complete DMEM growth medium (48 h), functional studies and parallel analysis of knockdown efficiency after cell lysis. The 4T1 cells with stable downregulation of E-cadherin (4T1-shCDH1) were generated using MISSION lentiviral particles containing the pLKO.1 construct to target mouse CDH1 (Sigma-Aldrich) and five different clones using different sequences were obtained (Supplementary Table 2). Control cells (4T1-shNT) were generated to express non-targeting sequence (Supplementary Table 2). MCF-7 cells were transfected with pSUPER-Ecadherin-shRNA (MCF-7-shCDH1) or pSUPER-Control-shRNA (MCF-7-shNT) using Lipofectamine RNAiMAX (ThermoFisher) and stable clones were selected using G418 to stably knock down E-cadherin, as described7. Stability of E-cadherin knockdown was verified by western blot and flow cytometry.
Scanning electron microscopy
Collagen gels were fixed with 2% glutaraldehyde in 0.1 M cacodylate buffer (1 h, 37 °C), washed (0.1 M cacodylate buffer), additionally fixed in 1% OsO4 in 0.1 M cacodylate buffer (1 h, 37 °C), dehydrated and analysed by scanning electron microscopy (Zeiss Sigma 300 Carl Zeiss).
Generation of spheroids
Cancer cell spheroids were generated using the hanging drop method37. In brief, cells from subconfluent culture were detached using 1 mM EDTA (ThermoFisher) and 0.075% trypsin (ThermoFisher), suspended in growth medium supplemented with 0.12% methylcellulose (Sigma-Aldrich) and maintained overnight as hanging droplets each containing 500–1,000 cells. For spheroid preparation with 4T1-shNT and 4T1-shCDH1 cells, pepsinized collagen type I from bovine skin (final concentration 10 μg ml−1, PureCol, Advanced BioMatrix) was additionally added to enhance cell aggregation.
Collagen 3D and interface migration assays
For 3D culture, spheroids were incorporated into pepsinized collagen type I from bovine skin (final concentration 1.6 mg ml−1, PureCol, Advanced BioMatrix) or non-pepsinized collagen from rat-tail tendon (final concentration between 2 mg ml−1 and 6 mg ml−1; BD Biosciences) before polymerization. Fibrillar collagen matrices containing spheroids were polymerized at 37 °C at pH 7.4 in customized glass chambers30 or mounted as drop-gels in 12-well plates and maintained at 37 °C. For cell migration along the interface between 3D fibrillar collagen and plastic, spheroids were allowed to sink to the bottom of the well before polymerization was complete. Cell migration from spheroids was monitored by bright-field microscopy using a CCD camera (Sentech), a ×10/0.20 numerical aperture (NA) air objective and frame-grabber system (Vistek) with a frame interval of 4 min or by end-point analysis after 24–72 h (4T1 cells) or 7 d (MCF-7 cells in coculture with fibroblasts).
Two-photon laser micropatterning
Multiphoton excitation at 830 nm (Coherent, Chameleon-Ultra I) was used to generate linear microtracks in pre-formed 3D rat-tail collagen matrices (6 mg ml−1), as described37. Tracks of 150 × 30 × 30 μm were positioned directly adjacent to the spheroid border and invasion was monitored for 24–48 h.
Immunofluorescence staining and confocal microscopy
Collagen lattices were fixed with 4% paraformaldehyde (30 min, 20 °C), incubated in PBS/10% goat serum/0.05% Triton X-100 (1 h, 20 °C), followed by incubation with primary antibody in PBS/0.1% BSA/0.05% Triton X-100 (4 °C, 20 °C), washed, and incubated with secondary antibody, DAPI and/or phalloidin (4 °C, 20 °C). Confocal fluorescence and reflection microscopy were performed using ×20/0.50 NA and ×40/0.80 NA objectives (Olympus FV1000).
Function inhibition assays
Interference in live-cell assays was performed using antibody against E-cadherin (clone SHE78–7) or isotypic control (IgG2a) at a final concentration of 10 μg ml−1. MMP activity was inhibited using the broad-spectrum inhibitor GM6001 (Ilomastat, 20 μM; Merck Millipore) or DMSO as a control. Antibodies, inhibitors and respective controls were added to the cell–collagen mixture before polymerization and to growth medium after collagen polymerization.
Immunohistochemistry
Formalin-fixed paraffin-embedded tissues from murine or human tumour lesions were sectioned into 4–5 μm slices (Microm HM340E, ThermoScientific), mounted on slides, air-dried (37 °C, 18 h) and deparaffinized using xylene and descending ethanol concentrations (100%–50%). Antigen retrieval was performed by boiling in Tris-EDTA buffer (pH 9.0, 15 min) followed by blocking of endogenous peroxidase activity in 3% hydrogen peroxide, washing (Tris-HCl, pH 7.5) and incubation with primary antibody (4 °C, 18 h). Samples were washed (Tris-HCl, pH 7.5), incubated with secondary goat anti-rabbit IgGs conjugated to a polymer labelled with horseradish peroxidase (DAKO, 30 min, room temperature) followed by incubation with 3,3′- diaminobenzidine (Sigma-Aldrich; 10 min, 20 °C) and haematoxylin (30 s) according to manufacturer’s protocol. Sections were mounted in Pertex medium (Histolab) and scanned (Pannoramic 250 Flash II, 3DHistech).
3D reconstruction of carcinoma lesions ex vivo
Tissue samples were obtained after surgical removal of primary breast cancer from seven patients with IDC and three patients with ILC (Department of Pathology, Radboud University Medical Center). From ten breast resection specimens, a representative tumour sample was taken (not influencing primary diagnostics; ‘leftover material’), consisting of seven cases of IDC and three cases of ILC. In addition, from these ten cases, a macroscopically normal-appearing breast tissue sample was taken at a distance from the primary tumour. Tumour samples were encrypted and analysed in an anonymized manner, as approved by the institutional review board and according to national law62, and in compliance with the Declaration of Helsinki. According to Dutch legislation, no approval from a research ethics committee was required for this study, as coded tissue obtained from routine diagnostic workflow was used and the included patients are not affected by the study. Anonymous or coded use of redundant tissue for research purposes is part of the standard treatment agreement with patients in the Radboud University Medical Center, to which patients may opt out. None of the included patients submitted an objection against use of residual material.
Tumour samples were fixed in formalin and sectioned with a vibratome (Leica, VT1000s) to obtain 200-μm-thick slices. For optimized antibody penetration, sequential 24 h incubations (4 °C) were performed for each primary and secondary antibody incubation and washing steps (PBS, 2% goat serum, 0.1% Triton X-100, 0.05% NaN3). Images were acquired using two-photon microscopy (LaVision BioTec, Miltenyi Biotec) with a ×20/0.95 NA objective (Olympus XLUMPlanFI 20x) and excitation at 910, 1,090 and 1,180 nm. Tumour samples obtained from murine models were prepared in the same way.
Flow cytometry
For surface-protein expression analysis, cells were detached with EDTA (2 mM), washed, incubated with primary antibody (10 μg ml−1 in PBS), washed, incubated with secondary Alexa Fluor 488-conjugated antibody (1:400) and analysed by flow cytometry (FACSCalibur, BD Biosciences) and FCS Express 3 software (De Novo Software).
SDS–PAGE and western blotting
Cells were lysed using SDS sample buffer (62.5 mM Tris-HCl, 2% SDS, 10% glycerol, 50 mM DTT and bromophenol blue). Cell lysates were loaded onto 6–15% acrylamide gels, followed by electrophoresis and wet or semi-dry transfer (iBlot, ThermoScientific) onto PVDF membranes (0.45 μm, Amersham Hybond), followed by fluorescence detection and the analysis of band intensities (Odyssey, LI-COR Biosciences).
Intravital two-photon microscopy
BALB/c (for 4T1 tumours) female mice (6–8 weeks old) were obtained from Charles River Laboratories. Animal procedures were approved by the Animal Ethics Committee of the Radboud University, Nijmegen (RU-DEC 2012–129) and performed according to the guidelines of the Dutch Act on Animal Experimentation and the European FELASA protocol. For implantation of the mammary window, the fourth mammary fat pad in anaesthetized mice (1–2% isoflurane in O2) was exposed by surgical incision under a Leica MZFLIII microscope. A superficial microchannel was created within the fat pad using a 30G syringe needle (BD Medical) into which an individual spheroid in a microdrop (2 μl in PBS) was implanted using a gel-loading pipette tip (0.3 mm inner diameter, BIOplastics) as described28. After implantation of up to eight spheroids in the same tissue area a mammary imaging window was inserted, affixed to the skin by gentle tightening of a purse-string suture around the skin rim, and closed with a cover-glass and spring ring, as described63. Intravital imaging was performed on a customized upright TrimScope II multiphoton microscope (LaVision BioTec, Miltenyi Biotec) equipped with three tunable Ti:Sa lasers (Coherent, Ultra I and II) and an Optical Parametric Oscillator (Coherent/APE) using a ×20 Olympus XLUMPlanFI ×20/0.95 NA water-immersion objective with a custom-made objective heater (37 °C) and up to 5 photomultipliers (Hamamatsu, Alkali H6780–01, H6780–20 or GaAsP H7422A-40) with the following filter configuration: 395/11 nm (third harmonic generation), 447/60 nm (DAPI), 525/50 nm (eGFP), 593/11 nm (second harmonic generation), 593/40 nm or 620/60 nm (mCherry); 675/67 nm (Alexa Fluor 647) (Semrock or Chroma Technology). The mammary imaging window of mice anaesthetized with 1–2% isoflurane in O2 was stabilized by a custom holder on the temperature-controlled stage (37 °C). 3D stacks were obtained with a 5 μm z-depth.
Spontaneous metastasis analysis
Control (shNT) or knockdown (shCDH1) MCF-7 cells (1 × 106 in 50 μl PBS with 1 mg ml−1 Matrigel) or 4T1 cells (1 × 105 in 50 μl PBS) were injected into the fourth mammary fat pad of imaging window-free BALB/c-nu/nu and BALB/c 8-week old female mice (Charles River Laboratories), respectively. Tumour growth was measured weekly with a caliper and mice were euthanized 30–36 d after cancer cell injection or when a tumour volume of 2 cm3 was reached. Lungs were collected, snap-frozen in liquid nitrogen, fixed in buffered formalin (4%, 5 d, 4 °C) followed by embedding in paraffin and sectioning (5-μm-thick slices of entire lungs). Because fluorescent reporter expression in metastatic sites of 4T1 tumours in vivo was not reliable, causing false-negative events, immunohistochemistry of cytokeratin-8 was used for scoring metastasis, based on reliable cytokeratin-8 expression after downregulation of E-cadherin in 4T1 cells (Extended Data Fig. 7b) and quantification was performed as described28. Cytokeratin-8-positive normal lung cells (pneumocytes) were excluded from quantification based on non-clustered cell morphology, position in alveoli and smaller size and regular shape of the nucleus compared with tumour cells. Metastasis was quantified as the number of neoplastic events, obtained from sequential sections with 70 μm intervals through the entire lung, and represented as the number of metastatic foci per slice. As minimum size for inclusion in the analysis, ≥ 3 cells were considered as metastatic cluster. To identify control (shNT) or knockdown (shCDH1) MCF-7 cells in the lungs, anti-E-cadherin or human-specific anti-vimentin antibodies were used, respectively (Extended Data Fig. 7c). No MCF-7 clusters or single cells were detected in the lungs for any condition.
Image analysis
The length of collectively invading strands, the area of sheet-like migration and the number of detached individual cells were quantified as the 2D area of the invasion region from projected 3D image stacks or from bright-field images, using manual image segmentation and analysis (ImageJ v.1.49s64). Images were processed by adjustment of brightness and contrast, cropping, rotation, segmentation, 3D reconstruction, mosaic stitching and drift correction during time-lapse recording. For measuring cell protein levels (E-cadherin or β-catenin), regions of interest were manually defined on multichannel overlay images around single cells (for cytoplasmic and membrane areas), and relative signal intensities were acquired.
In silico model of migration plasticity
A lattice-gas cellular automaton model44,45 was used to test the effects of E-cadherin downregulation on collective and single-cell migration modes under conditions of different ECM confinement. The detailed mathematical procedures, the simulation code and videos of example realizations were deposited at GitHub (https://github.com/sisyga/jamminglgca). Cell dynamics were determined by a stochastic process depending on the local, spatial microenvironment defined by the cell and ECM density (Fig. 5a). Cells were represented by point particles with discrete velocities moving between adjacent sites on a regular lattice with a lattice spacing of 20 μm in discrete 15-min time steps. The ECM was represented by a scalar field defined on the lattice to reflect the local level of confinement. We assumed that confinement by the ECM reduces cell speed, which we implemented by increasing the probability of cells switching to the resting state. Cadherin-mediated adhesive interactions between cells were integrated as a tendency of cells to move to and stay in the neighbourhood of other cells, and by increased cohesion between cells, leading to alignment of cell velocities. We assumed that the invading cells can open the gap between the collagen-coated plastic surface and the overlaying collagen network (Fig. 5b, cartoon). This is implemented by removal of the ECM component by the invading cells. We idealized the interface invasion assay by monitoring the invasion into a two-dimensional hexagonal lattice under conditions of varying ECM density and cell–cell adhesion strength. As the initial condition, a line of cells was placed at the lower (y = 0) line of the lattice at high confluency. We then monitored the movement along the y direction. Cells were supplemented at the y = 0 border at constant rate (r) = 0.05 per time step, representing a constant influx of cells from the tumour spheroid. The evolution of the system was equilibrated over an initial phase of 50 time steps, to ensure a quasi-steady state, and then monitored for 200 time steps corresponding to approximately 48 h of observation time. To characterize the mode of migration, we selected two observables: the number of single cells and the velocity correlation between next neighbours. Here, we regarded cells without next neighbours, that is, no other cells on the same or next-neighbour lattice sites as single cells. For each parameter combination, we determined the accumulated number of single cells and the next-neighbour velocity correlations as the average of 5 independent simulations and 200 time steps.
Cell tracking
Cells were reliably detected and tracked using fluorescently labelled nuclei (H2B–mCherry) (ImageJ or Fiji64,65, plugin TrackMate66) by applying a spot size of 10 μm and a threshold of 0.5. The detected spots were linked from frame to frame using the TrackMate LAP Tracker, penalizing inter-spot differences in median spot intensity, standard deviation of intensity and estimated spot diameter. This enabled cell tracking even when the cells closely passed by each other, and avoided track ‘jumps’ between cells. The maximum allowed linking distance was adjusted specifically for the different cell type and collagen concentration, such that the maximally allowed step size was slightly larger than the corresponding maximal frame-to-frame velocities. The quality of the resulting tracks was verified by visual inspection for all combinations of cell types and collagen concentrations. Tracks shorter than ten frames were excluded from analysis. This analysis was used for both the 2D in vitro trajectories and the 3D in vivo trajectories. The 3D data was registered using a custom MATLAB script before tracking (https://github.com/steffengrosser/smart-3D-registration-2019). The correlation analysis of the tracks was calculated using a custom MATLAB algorithm (https://github.com/JuergenLippoldt/On-Motility-and-Form).
The core region was defined as the track within a 100 μm radius of the spheroid centre, and the edge was defined as the 10% outermost cells. All other regions were considered as the mid zone. For the spatial and temporal correlation functions Pearson correlation coefficients were used. Individual velocity correlations were mapped over the distance between the compared cells and over the lag time for the temporal correlation function and averaged with a Gaussian kernel. For the mean squared displacement of in vitro experiments the average squared displacements were calculated from cells within the respective regions, and for in vivo experiments the data were averaged over the experiment.
Cell segmentation
Two-dimensional cell-shape approximations were obtained on the basis of the boundary information from the phalloidin channel and the cell-centre information from the nucleus, using a custom MATLAB cell segmentation script (https://github.com/JuergenLippoldt/On-Motility-and-Form) for confocal slides with stained nuclei and F-actin. In brief, the actin signal was used in a watershed algorithm using markers of nuclei and cell-free regions. The nuclei were detected using adaptive thresholds and separated using maxima of a smoothed distance to the closest non-thresholded pixel. To test this method, we compared manually corrected segmentations to the results from this fully automatic algorithm. The differences between the cell shape distributions of the manually corrected and the automatic segmentations were much smaller than between intervention groups.
Quantification of velocities and vorticities by particle image velocimetry
PIV was applied to fluorescent images of nuclei in various conditions, using the PIVLab67 application for MATLAB. The method estimates the local velocities by digital image correlation. To reduce non-stationary effects due to the expansion of the spheroid, the analysis was limited to 20 subsequent frames starting after t = 1,050 min (frame interval: 15 min). Background noise was effectively filtered by a median filter using the medfilt2 (https://de.mathworks.com/help/images/ref/medfilt2.html) function (MATLAB), intensity thresholding and, on the basis of intensity histograms, thresholding (value of 10). The velocity field was obtained as pixels per frame and converted to μm h−1. The vorticity of the velocity field was computed using the standard MATLAB function curl (https://de.mathworks.com/help/symbolic/curl.html).
To obtain zone-related data, regions of interest within 50 concentric rings centred on the spheroid were analysed (Fig. 6i). Data from three larger concentric regions of interest were pooled, denoting as core, bulk, and edge, based on their distance R to the spheroid center: , for core; , for bulk and for edge. Here, is the core radius, and are the inner and outer radii of the bulk region, and are the inner and outer radii of the edge region. Due to differences in migration speed the regions were adapted to 2 mg ml−1 and 6 mg ml−1 collagen conditions (Supplementary Table 3). Data were pooled from two independent experiments, each consisting of five replicates per condition, and the means were averaged. Statistical significance was evaluated using the Kolmogorov–Smirnov test with a Bonferroni correction for multiple comparisons.
Statistics and reproducibility
Statistical analysis was performed as the non-parametric two-tailed Mann–Whitney test; a Student’s t-test using Fisher z-transformation or Kolmogorov–Smirnov test (using the ks_2samp function of the SciPy Python library) with or without a Bonferroni correction for multiple comparisons. Unless stated otherwise, all results were reproduced by at least three independent experiments. No datasets were excluded from data display and analysis. All data replicated in independent experiments were reproducible. Further details about the replication of data are stated in the figure legends. The data represented in the figures were from one (Fig. 2f) or two (Extended Data Figs. 3e,g, 4g, 6a–c and 7a) independent experiments. Multiple independent spheroids were pooled from one (Fig. 6c,d) or two (Figs. 3g, right, 6b,e–h,j,k and Extended Data Fig. 6d–g) experimental cohorts.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that data supporting the findings of this study are available within the paper (and its supplementary information files), and are available from the corresponding author on reasonable request. Source data are provided with this paper.
Code availability
The detailed mathematical procedures, the simulation code and videos of example realizations for in silico modelling were deposited at GitHub (https://github.com/sisyga/jamminglgca). Custom MATLAB scripts for cell-nuclei tracking and cell segmentation are available at https://github.com/steffengrosser/smart-3D-registration-2019 and https://github.com/JuergenLippoldt/On-Motility-and-Form. Codes for the PIV analysis are available at https://github.com/ComplexityBiosystems/PIV-spheroids.
Extended Data
Extended Data Fig. 1. 3D organization of normal and mammary carcinoma tissue and the geometry of interstitial tissue tracks.
a, Immunofluorescence staining of E-cadherin or pan-cytokeratin (pan-CK), vimentin and collagen IV in normal ducts, IDC and ILC lesions. Arrowheads indicate single carcinoma cells. Example images represent 3 patient samples per condition. b, 3D reconstruction of z-projection (200 μm thickness) from IDC and ILC samples with carcinoma cells oriented in the niches formed by aligned collagen fibers. Example images represent 7 (IDC) and 3 (ILC) patient samples. c, 3D rendering of collective carcinoma strands and tissue tracks in IDC (left panel), and a different region with tissue tracks formed by collagen bundles (right panel). d, Tissue structure surrounding E-cadherin positive duct in normal breast tissue. Example image represents 3 normal breast tissue patient samples. e, Inter-fiber distance of tissue tracks formed by collagen bundles with stroma cells present or absent within the tracks. Data represent n = 49 and 24, 139 and 78, and 107 and 93 cell-free or cell-filled tissue microtracks pooled from 3 (normal breast tissue), 7 (tumor-free IDC tissue) and 3 (tumor-free ILC tissue) patient samples, respectively. Median (red line) and dots representing individual tissue microtracks. P values, two-tailed Mann-Whitney test. Scale bars, 50 μm.
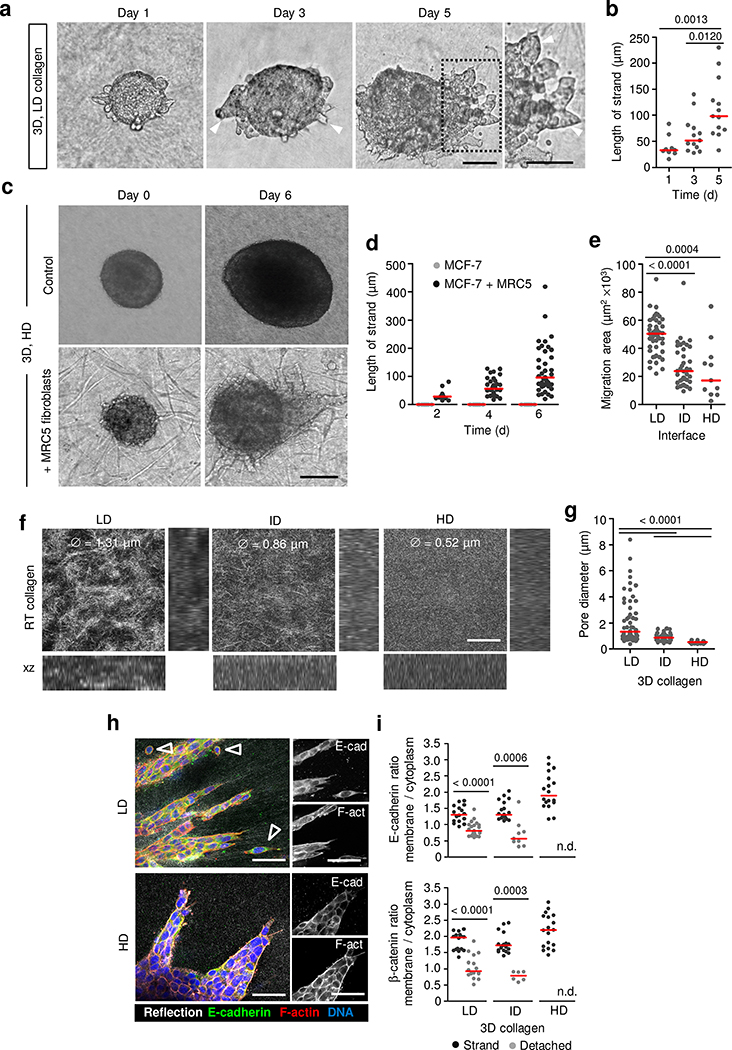
Extended Data Fig. 2. Collective invasion of breast cancer cells in 3D collagen matrix culture enriched with fibroblasts or fibroblast-free matrix of different density.
a, Time-dependent collective invasion of MCF-7 spheroid into LD collagen matrix. Micrographs represent 3 independent experiments. b, Collective strand length. Data represent average length of strands from n = 9 (day 1), 13 (day 3) and 13 (day 5) spheroids (3 independent experiments). c, MCF-7 cell invasion into HD collagen matrix in dependence of cocultured MRC-5 fibroblasts. Micrographs represent 3 independent experiments. d, Length of invasion strands with and without cocultured MRC-5 fibroblasts. Data without MRC-5 represent n = 15 spheroids (day 2, 4, 6) and with MRC-5, n = 13 (day 2), 31 (day 4), 39 (day 6) spheroids/condition (3 independent experiments). e, Area of collective MCF-7 sheet moving along the collagen interface of different density. Data represent n = 47 (LD), 38 (ID), 11 (HD) spheroids/condition (3 independent experiments). f, Rat-tail (RT) collagen network at 2 (LD), 4 (ID) and 6 mg/mL (HD) density, used for 3D invasion of 4T1 cells. g, Pore diameters at different collagen densities. Data represent n = 56 (LD), 41 (ID), 26 (HD) pores/condition (3 independent experiments). h, E-cadherin localization in 4T1 cells during collective invasion in collagen with different density (48 h). Micrographs represent 3 independent experiments. i, Cell boundary/cytoplasm ratio of E-cadherin and β-catenin intensity (conditions as in h). n.d., no detached cells detectable. Data for E-cadherin represent n = 16, 19 and 18 (strands) and 21 and 9 (detached) cells for LD, ID and HD conditions, respectively, and for β-catenin 16, 19 and 18 (strands) and 16 and 6 (detached) cells for LD, ID and HD conditions, respectively (3 independent experiments). Medians (red line); dots, means from individual spheroids (b, d, e), collagen pores (g) or individual cells (i). P values, two-tailed Mann-Whitney test. Collagen matrices described in Supplementary Table 1. Scale bars, 200 μm (a, c); 50 μm (f, h).
Extended Data Fig. 3. Collective invasion of MCF-7 cells in dense collagen matrix depends on MMP activity.
a, E-cadherin localization in collectively invading MCF-7 cells in coculture with MRC-5 fibroblasts. Micrograph representing 3 independent experiments. b, Fibroblast-generated tracks of least resistance in HD collagen containing MCF-7 strand revealed by Col 3/4 staining. Micrograph from 3 independent experiments. c, MRC-5 and HMF fibroblasts-supported invasion of MCF-7 cells into HD collagen. Micrograph from 3 independent experiments. d, MCF-7 strand length in coculture with MRC-5 and HMF fibroblasts. Median (red line); dots represent n = 34 (MRC-5) and 27 (HMF) spheroids/condition (3 independent experiments). e, MT1-MMP/MMP14 expression by MRC-5, HMF and MCF-7 cells. Western blot from 2 independent experiments. Unmodified scan of the Western blot is provided in Source data. f, Collective invasion of MCF-7 cells into HD collagen matrix after overexpression of MT1-MMP/MMP14. Insert, parental MCF-7 cell spheroid. g, Number of collective MCF-7 strands in HD collagen and MCF-7 cells after MT1-MMP overexpression. Data represent n = 17 (parental) and 30 (MMP14-OE) spheroids/condition (2 independent experiments). h-j, MCF-7 collective invasion into HD collagen with fibroblasts (h), LD collagen (i), and LD or ID interface assay (j) after interference with MMPs (GM6001). Micrographs in (h-j) from 3 independent experiments. Length of MCF-7 strands (h, i) or sheet area (j). Data in (h-i) represent n = 71/71 (h), 44/39 (i) spheroids for DMSO/GM6001, respectively, or in (j) n = 21/32 (LD) and 16/10 (ID) spheroids for DMSO/GM6001, respectively (3 independent experiments). Median (red line), dots representing the means/spheroid (d, g, h, i, j). P values, two-tailed Mann-Whitney test. Low (LD), intermediate (ID) and high (HD) density collagen matrices are described in Supplementary Table 1. Scale bars, 100 μm (a, b); 200 μm (c, f, h-j).
Extended Data Fig. 4. E-cadherin downregulation in MCF-7 and 4T1 cells – impact on collective invasion in collagen matrix.
a-f, Expression of E-cadherin in MCF-7-shCDH1 (a-c) and 4T1-shCDH1 cells (d-f). Example forward and side scatter plot (FSC/SSC; a, d) for gating of morphologically intact cells (3 independent experiments). Relative fluorescence intensity in MCF-7-shCDH1 cells (b) and 4T1 cells after E-cadherin downregulation with 5 different shRNA sequences (e). Western blots in (c, f) represent 3 independent experiments. Unmodified scan of the Western blots is provided in Source data. g, Expression of adhesion molecules and EMT markers after shRNA-mediated downregulation of E-cadherin in MCF-7 and 4T1 cells. Western blots represent 2 independent experiments. Unmodified scan of the Western blots is provided in Source data. h, Number of collective strands in 4T1 spheroids in matrix with different collagen densities. Dots represent means from n = 12/10 (LD), 14/11 (ID), 15/7 (HD) spheroids for shNT/shCDH1, respectively (3 independent experiments). i, Length of 4T1 collective strands in 4T1 spheroids in different collagen density. Dots represent n = 130/10 (LD), 161/26 (ID), 107/19 (HD) strands for shNT/shCDH1, respectively (3 independent experiments). j, Collective invasion of MCF-7 cells after E-cadherin downregulation during coculture with HMF fibroblasts in HD collagen. Micrographs represent 3 independent experiments. k, MCF-7 strand length in the absence or presence of HMF fibroblasts. Dots represent the average strand length per spheroid from n = 30/29 (shNT) or 20/44 (shCDH1) spheroids for either HMF condition, respectively (3 independent experiments). l, E-cadherin surface localization in MCF-7-shNT or MCF-7-CDH1 cells migrating in LD or HD interface assay. Micrographs represent 3 independent experiments. Medians (red line) in (h, i, k). P-values, two-tailed Mann-Whitney test. Low (LD), intermediate (ID) and high (HD) density collagen matrices are described in Supplementary Table 1. Scale bars, 100 μm (j); 50 μm (l).
Extended Data Fig. 5. Collagen matrix confinement determines invasion mode of breast cancer cells in the presence of adhesion-perturbing anti-E-cadherin antibody.
a, Single-cell dissemination of MCF-7 cells in collagen matrix of different density in the absence or presence of blocking anti-E-cadherin mAb SHE78–7. Micrographs represent 3 independent experiments. b, Number of individualized cells under conditions in (a). Dots for 3D represent n = 11/16 (LD), 11/10 (ID), 34/34 (HD) and, for interface, n = 39/36 (LD), 22/16 (ID), 50/39 (HD) spheroids for IgG/SHE78–7, respectively, from 3 independent experiments. c, Dependence of the number of individually invading cells out of MCF-7 spheroids on the pore diameter within 3D collagen matrices with different density, with culture conditions as in (b). d, Single-cell detachment of MCF-7 cells by SHE78–7 mAb in LD collagen. Micrograph represents 3 independent experiments. e, Single-cell detachment of MCF-7 cells along collagen-ablated tracks in HD collagen. Micrograph represents 3 independent experiments. f, Number of individualized MCF-7 cells in collagen-ablated tracks. After 48 h tracks were completely filled with cells, resulting in multicellular strands irrespective of antibody condition. Data represent n = 7/18 (24 h), 7/18 (48 h) spheroids for IgG/SHE78–7, respectively pooled from 3 independent experiments. g, Combined interference using antibody SHE78–7 in MCF-7-shCDH1 cells migrating in interface assay with collagen of different density. Micrographs represent 3 independent experiments. h, Number of individualized MCF-7 cells after E-cadherin downregulation in interface assay. Data represent the means from n=20 spheroids per condition pooled from 3 independent experiments. Red line in (b, f, h), median. P values, two-tailed Mann-Whitney test. Low (LD), intermediate (ID) and high (HD) density collagen matrices are described in Supplementary Table 1. Scale bars, 100 μm (a, g); 50 μm (d, e).
Extended Data Fig. 6. Variability of cell-cell junction composition and kinetics of carcinoma cell invasion in dependence of E-cadherin status and ECM confinement.
a, b, c, F-actin (a, b), E-cadherin and p120 catenin (c) localization in 4T1-shNT or 4T1-shCDH1 cells invading from multicellular spheroids along the interface of LD (a) or HD (b, c) collagen after 48 h. Micrographs in (a-c) represent cells from 2 independent experiments. Scale bars, 50 μm. d-f, Automated tracking of H2B-mCherry positive cell nuclei of 4T1 cells moving along the interface of LD or HD collagen (time period: 27–54 h). Spatial correlation of velocity (d), temporal correlation of velocity (e) and mean squared displacement (f) of 4T1-shNT or 4T1-shCDH1 cells at the outward edge. Lines in (d-f) represent the average of n=10 spheroids/condition pooled from 2 independent experiments. g, Particle image velocimetry (PIV) analysis of H2B-mCherry positive cell nuclei of 4T1 cells in LD or HD interface assay (time period: 43–48 h). Vorticities and velocities obtained from 4T1 cell nuclei at the invasion edge, mid-invasion zone or in the spheroid core. Data represent n=10 spheroids/condition pooled from 2 independent experiments. Box plots in (g) represent the median (mid-line), the second quartile (colored band) and the 1.5 interquartile range (whiskers). Two-sided Kolmogorov-Smirnov tests indicate that all possible comparisons are statistically significant (p < 10−6).
Extended Data Fig. 7. Effects of E-cadherin downregulation on protein expression, tumor growth and spontaneous metastasis of MCF-7 and 4T1 cells after implantation into the mammary fat pad.
a, Collective invasion of 4T1 cells into the fibroblast-rich stroma 4 days after spheroid implantation into the mammary fat pad. Example images from 2 independent tumors. b, Pan-cytokeratin (upper panels) or cytokeratin-8 (lower panels) expression in 4T1-shNT or 4T1-shCDH1 tumors in the mammary fat pad (4 days post-implantation) or in the lungs (38 days after tumor implantation in the mammary fat pad). Micrographs represent 3 independent tumor samples (upper panel) or lungs samples from 5 mice/condition (lower panels). c, Pan-cytokeratin (upper panels) and E-cadherin or vimentin (lower panels) expression in MCF-7-shNT or MCF-7-shCDH1 tumors in the mammary fat pad (6 days post-implantation). Micrographs represent 2 (upper panel) or 1 (lower panels) tumors/condition. d, Time-dependent growth in MCF-7-shNT or MCF-7-shCDH1 primary tumors. Data display the mean ± SEM from n=5 mice per group. P values, two-tailed Mann-Whitney test. e, Number of metastatic foci within the lungs of MCF-7-shNT or MCF-7-shCDH1 tumor-bearing mice up to 38 days post-implantation in the mammary fat pad. Data represent the number of metastatic cell clusters per tissue slice (from 50–100 slices per lung) from n=5 (4T1-shNT, 4T1-shCDH1, MCF-7-shCDH1) or n=6 (MCF-7-shNT) mice/group. Red line, median. P values, two-tailed Mann-Whitney test. Scale bars, 50 μm (a, b, upper panel, c) and 25 μm (b, lower panel). f, Hypothetical jamming phase diagram for cell invasion into 3D tissue.
Supplementary Material
Supplementary Video 1
3D reconstruction of invasive ductal carcinoma lesion in a patient sample (30 μm z-stack, 5 μm step). Collective strands of carcinoma cells were detected by positive E-cadherin staining; collagen bundles (SHG); nuclei (DAPI). The video is representative for 7 independent samples. Related to Fig. 1.
Supplementary Video 2
3D reconstruction of invasive lobular carcinoma lesion in a patient sample (60 μm z-stack, 5 μm step). Collective strands of carcinoma cells were detected by positive staining for pan-cytokeratin; collagen bundles (SHG); nuclei (DAPI). The video is representative for 3 independent samples. Related to Fig. 1
Supplementary Video 3
In silico analysis of collective and single-cell migration modes in dependence of adhesion and extracellular matrix confinement. Time evolution of representative examples of four parameter combinations corresponding to low/high cell-cell adhesion and low/high ECM confinement. Strong cell-cell adhesion leads to correlated movement without single cell release (lower row, corresponding to Fig. 5e region 2); decreasing cell-cell adhesion leads to uncorrelated movement with single cell release if ECM confinement is weak (top left, corresponding to Fig. 5e region 1) and without single cell release if confinement is high (top right, corresponding to Fig. 5e region 3). Vectors correspond to local flux of cells. The color code indicates the local velocity correlation between nearest neighbours, with red/green corresponding to anti-correlated/correlated movement. Blue circles, single cells.
Supplementary Video 4
Time-lapse movies of the invasion of control (shNT) or shCDH1 4T1 cells from multicellular spheroids along the interface underneath the 3D collagen matrix of 2 mg/ml concentration. Bright-field and H2B-mCherry (nuclei) imaging. Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 5
Time-lapse movies of the invasion of control (shNT) or shCDH1 4T1 cells from multicellular spheroids along the interface underneath the 3D collagen matrix of 6 mg/ml concentration. Bright-field and H2B-mCherry (nuclei) imaging. Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 6
Automatic tracking of H2B-mCherry positive nuclei (magenta circles) of control (shNT) or shCDH1 4T1 cells invading from multicellular spheroids along the interface underneath the 3D collagen matrix of 2 mg/ml or 6 mg/ml concentration. Migration tracks (yellow lines). Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 7
PIV analysis of time lapse movies. The invasion of control (shNT) or shCDH1 4T1 cells from multicellular spheroids along the interface underneath the 3D collagen matrix of 2 mg/ml or 6 mg/ml concentration. The color denotes the vorticity and the arrows indicate the direction and amplitude of the local velocity. Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 8
Automatic 3D tracking of H2B-mCherry/LifeAct-eGFP shNT or shCDH1 4T1 cells in vivo 4 days after multicellular spheroid implantation in the mouse mammary fat pad. Time-lapse movie representing the maximum projection of 85 μm Z-stack taken every 10 min over 140 min. Related to Fig. 7. The video is representative for 3 independent experiments per condition.
Acknowledgements
We gratefully acknowledge the Microscopical Imaging Centre core support by H. Croes and J. Fransen; M. K. te Lindert for scanning electron microscopy; B. Lemmers-Van de Weem for the excellent technical assistance in animal experiments; and L. Wolf for 3D rendering of carcinoma invasion in patient samples. We thank the Centre for Information Services and High Performance Computing at TU Dresden for providing computational infrastructure. This work was supported by Deutsche Krebshilfe (106950 and 70112014), the European Union FP7 Tissue Transmigration Training Network (T3Net 237946), the Netherlands Science Organization (NWO-VICI 918.11.626), the European Research Council (617430-DEEPINSIGHT), NIH-U54 CA210184-01, and the Cancer Genomics Center (CGC.nl) to P.F.; the German Research Foundation (Deutsche Forschungsgemeinschaft, SFB-TR 79) to A.D.; the European Social Fund (ESF), co-financed by tax funds on the basis of the budget adopted by the members of the Saxon State Parliament, to S.S.; the European Research Council (ERC-741350) to J.A.K.; AIRC under IG2018-ID 21558 (principal investigator, M. Pusch) to C.A.M.L.P.; and the Lise–Meitner fellowship M 2450-NBL by the Austrian Science Fund (FWF) to O.C.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41556-020-0552-6.
Supplementary information is available for this paper at https://doi.org/10.1038/s41556-020-0552-6
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Mayor R & Etienne-Manneville S The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol 17, 97–109 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Friedl P, Locker J, Sahai E & Segall JE Classifying collective cancer cell invasion. Nat. Cell Biol 14, 777–783 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Gomez GA, McLachlan RW & Yap AS Productive tension: force-sensing and homeostasis of cell–cell junctions. Trends Cell Biol. 21, 499–505 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Reffay M et al. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat. Cell Biol 16, 217–223 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Ewald AJ, Brenot A, Duong M, Chan BS & Werb Z Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell 14, 570–581 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sunyer R et al. Collective cell durotaxis emerges from long-range intercellular force transmission. Science 353, 1157–1161 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Lehembre F et al. NCAM-induced focal adhesion assembly: a functional switch upon loss of E-cadherin. EMBO J. 27, 2603–2615 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wendt MK, Taylor MA, Schiemann BJ & Schiemann WP Down-regulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol. Biol. Cell 22, 2423–2435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perl AK, Wilgenbus P, Dahl U, Semb H & Christofori G A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature 392, 190–193 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Ye X & Weinberg RA Epithelial–mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 25, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derksen PWB et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 10, 437–449 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Khalil AA et al. Collective invasion in ductal and lobular breast cancer associates with distant metastasis. Clin. Exp. Metastasis 34, 421–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalski PJ, Rubin MA & Kleer CG E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 5, R217–R222 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narendra S, Jenkins SM, Khoor A & Nassar A Clinical outcome in pleomorphic lobular carcinoma: a case-control study with comparison to classic invasive lobular carcinoma. Ann. Diagn. Pathol 19, 64–69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padmanaban V et al. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573, 439–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beerling E et al. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 14, 2281–2288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer KR et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aceto N et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bi D, Yang X, Marchetti MC & Manning M Motility-driven glass and jamming transitions in biological tissues. Phys. Rev. X 6, 021011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haeger A, Krause M, Wolf K & Friedl P Cell jamming: collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim. Biophys. Acta 1840, 2386–2395 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Doxzen K et al. Guidance of collective cell migration by substrate geometry. Integr. Biol 5, 1026–1035 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Gaggioli C et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol 9, 1392–1400 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Weigelin B, Bakker G & Friedl P Intravital third harmonic generation microscopy of collective melanoma cell invasion. IntraVital 1, 32–43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Helvert S, Storm C & Friedl P Mechanoreciprocity in cell migration. Nat. Cell Biol 20, 8–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedl P & Mayor R Tuning collective cell migration by cell–cell junction regulation. Cold Spring Harb. Perspect. Biol 9, a029199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duclos G et al. Spontaneous shear flow in confined cellular nematics. Nat. Phys 14, 728–732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf K et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol 9, 893–904 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Ilina O et al. Intravital microscopy of collective invasion plasticity in breast cancer. Dis. Model Mech 11, dmm034330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Provenzano PP et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 6, 11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf K et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol 201, 1069–1084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szabo A et al. In vivo confinement promotes collective migration of neural crest cells. J. Cell Biol 213, 543–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balzer EM et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 26, 4045–4056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haeger A et al. Collective cancer invasion forms an integrin-dependent radioresistant niche. J. Exp. Med 217, e20181184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bi D, Lopez JH, Schwarz JM & Manning ML A density-independent rigidity transition in biological tissues. Nat. Phys 11, 1074 (2015). [Google Scholar]
- 35.Park J-A et al. Unjamming and cell shape in the asthmatic airwayepithelium. Nat. Mater 14, 1040–1048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palamidessi A et al. Unjamming overcomes kinetic and proliferation arrest in terminally differentiated cells and promotes collective motility of carcinoma. Nat. Mater 18, 1252–1263 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Ilina O, Bakker G-J, Vasaturo A, Hofmann RM & Friedl P Two-photon laser-generated microtracks in 3D collagen lattices: principles of MMP-dependent and -independent collective cancer cell invasion. Phys. Biol 8, 15010 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Singhai R et al. E-Cadherin as a diagnostic biomarker in breast cancer. N. Am. J. Med. Sci 3, 227–233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vos CB et al. E-cadherin inactivation in lobular carcinoma in situ of the breast: an early event in tumorigenesis. Br. J. Cancer 76, 1131–1133 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conklin MW et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol 178, 1221–1232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Provenzano PP et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4, 38 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elisha Y, Kalchenko V, Kuznetsov Y & Geiger B Dual role of E-cadherin in the regulation of invasive collective migration of mammary carcinoma cells. Sci. Rep 8, 4986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cadwell CM, Su W & Kowalczyk AP Cadherin tales: Regulation of cadherin function by endocytic membrane trafficking. Traffic 17, 1262–1271 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Frisch U, Hasslacher B & Pomeau Y Lattice-gas automata for the Navier–Stokes equation. Phys. Rev. Lett 56, 1505–1508 (1986). [DOI] [PubMed] [Google Scholar]
- 45.Deutsch A & Dormann S Cellular Automaton Modeling of Biological Pattern Formation (Birkhäuser, 2018). [Google Scholar]
- 46.Bussemaker HJ, Deutsch A & Geigant E Mean-field analysis of a dynamical phase transition in a cellular automaton model for collective motion. Phys. Rev. Lett 78, 5018–5021 (1997). [Google Scholar]
- 47.Deutsch A A new mechanism of aggregation in a lattice-gas cellular automaton model. Math. Comput. Modell 31, 35–40 (2000). [Google Scholar]
- 48.Duclos G, Garcia S, Yevick HG & Silberzan P Perfect nematic order in confined monolayers of spindle-shaped cells. Soft Matter 10, 2346–2353 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Schotz E-M, Lanio M, Talbot JA & Manning ML Glassy dynamics in three-dimensional embryonic tissues. J. R. Soc. Interface 10, 20130726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Notbohm J et al. Cellular contraction and polarization drive collective cellular motion. Biophys. J 110, 2729–2738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atia L et al. Geometric constraints during epithelial jamming. Nat. Phys 14, 613–620 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayer A et al. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat. Cell Biol 18, 1311–1323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bazellieres E et al. Control of cell–cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol 17, 409–420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Angelini TE et al. Glass-like dynamics of collective cell migration. Proc. Natl Acad. Sci. USA 108, 4714–4719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia S et al. Physics of active jamming during collective cellular motion in a monolayer. Proc. Natl. Acad. Sci. USA 112, 15314–15319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Douezan S et al. Spreading dynamics and wetting transition of cellular aggregates. Proc. Natl Acad. Sci. USA 108, 7315–7320 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Truong HH et al. β1 integrin inhibition elicits a prometastatic switch through the TGFβ-miR-200-ZEB network in E-cadherin-positive triple-negative breast cancer. Sci. Signal 7, ra15 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Shamir ER et al. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J. Cell Biol 204, 839–856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aiello NM et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell 45, 681–695 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuriyama S et al. In vivo collective cell migration requires an LPAR2-dependent increase in tissue fluidity. J. Cell Biol 206, 113–127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiesner C et al. KIF5B and KIF3A/KIF3B kinesins drive MT1–MMP surface exposure, CD44 shedding, and extracellular matrix degradation in primary macrophages. Blood 116, 1559–1569 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Nagelkerke A et al. Constitutive expression of γ-H2AX has prognostic relevance in triple negative breast cancer. Radiother. Oncol 101, 39–45 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Alieva M, Ritsma L, Giedt RJ, Weissleder R & van Rheenen J Imaging windows for long-term intravital imaging: general overview and technical insights. Intravital 3, e29917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rueden CT et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tinevez J-Y et al. TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Thielicke W & Stamhuis EJ PIVlab—towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw 2, e30 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1
3D reconstruction of invasive ductal carcinoma lesion in a patient sample (30 μm z-stack, 5 μm step). Collective strands of carcinoma cells were detected by positive E-cadherin staining; collagen bundles (SHG); nuclei (DAPI). The video is representative for 7 independent samples. Related to Fig. 1.
Supplementary Video 2
3D reconstruction of invasive lobular carcinoma lesion in a patient sample (60 μm z-stack, 5 μm step). Collective strands of carcinoma cells were detected by positive staining for pan-cytokeratin; collagen bundles (SHG); nuclei (DAPI). The video is representative for 3 independent samples. Related to Fig. 1
Supplementary Video 3
In silico analysis of collective and single-cell migration modes in dependence of adhesion and extracellular matrix confinement. Time evolution of representative examples of four parameter combinations corresponding to low/high cell-cell adhesion and low/high ECM confinement. Strong cell-cell adhesion leads to correlated movement without single cell release (lower row, corresponding to Fig. 5e region 2); decreasing cell-cell adhesion leads to uncorrelated movement with single cell release if ECM confinement is weak (top left, corresponding to Fig. 5e region 1) and without single cell release if confinement is high (top right, corresponding to Fig. 5e region 3). Vectors correspond to local flux of cells. The color code indicates the local velocity correlation between nearest neighbours, with red/green corresponding to anti-correlated/correlated movement. Blue circles, single cells.
Supplementary Video 4
Time-lapse movies of the invasion of control (shNT) or shCDH1 4T1 cells from multicellular spheroids along the interface underneath the 3D collagen matrix of 2 mg/ml concentration. Bright-field and H2B-mCherry (nuclei) imaging. Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 5
Time-lapse movies of the invasion of control (shNT) or shCDH1 4T1 cells from multicellular spheroids along the interface underneath the 3D collagen matrix of 6 mg/ml concentration. Bright-field and H2B-mCherry (nuclei) imaging. Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 6
Automatic tracking of H2B-mCherry positive nuclei (magenta circles) of control (shNT) or shCDH1 4T1 cells invading from multicellular spheroids along the interface underneath the 3D collagen matrix of 2 mg/ml or 6 mg/ml concentration. Migration tracks (yellow lines). Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 7
PIV analysis of time lapse movies. The invasion of control (shNT) or shCDH1 4T1 cells from multicellular spheroids along the interface underneath the 3D collagen matrix of 2 mg/ml or 6 mg/ml concentration. The color denotes the vorticity and the arrows indicate the direction and amplitude of the local velocity. Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 8
Automatic 3D tracking of H2B-mCherry/LifeAct-eGFP shNT or shCDH1 4T1 cells in vivo 4 days after multicellular spheroid implantation in the mouse mammary fat pad. Time-lapse movie representing the maximum projection of 85 μm Z-stack taken every 10 min over 140 min. Related to Fig. 7. The video is representative for 3 independent experiments per condition.
Data Availability Statement
The authors declare that data supporting the findings of this study are available within the paper (and its supplementary information files), and are available from the corresponding author on reasonable request. Source data are provided with this paper.