Significance
Genetic systems for controlling transmission of vector-borne diseases are moving from discovery-stage demonstrations of proofs-of-principle to the next phases of development. A successful transition requires meeting safety and efficacy criteria defined in target product profiles. We show here that the Cas9/guide RNA-based gene-drive components of a genetically-engineered malaria mosquito vector, Anopheles gambiae, achieve key target product profile requirements for efficacy and performance. This system is designed to achieve mosquito population modification when coupled with genes encoding antiparasite effector molecules and result in stable and sustainable blocking of malaria parasite transmission.
Keywords: cage trials, off-target, nontarget, load, guide RNA polymorphisms
Abstract
A Cas9/guide RNA-based gene drive strain, AgNosCd-1, was developed to deliver antiparasite effector molecules to the malaria vector mosquito, Anopheles gambiae. The drive system targets the cardinal gene ortholog producing a red-eye phenotype. Drive can achieve 98 to 100% in both sexes and full introduction was observed in small cage trials within 6 to 10 generations following a single release of gene-drive males. No genetic load resulting from the integrated transgenes impaired drive performance in the trials. Potential drive-resistant target-site alleles arise at a frequency <0.1, and five of the most prevalent polymorphisms in the guide RNA target site in collections of colonized and wild-derived African mosquitoes do not prevent cleavage in vitro by the Cas9/guide RNA complex. Only one predicted off-target site is cleavable in vitro, with negligible deletions observed in vivo. AgNosCd-1 meets key performance criteria of a target product profile and can be a valuable component of a field-ready strain for mosquito population modification to control malaria transmission.
Recent applications of Cas9/guide RNA (gRNA) technologies support the development of genetically-engineered mosquitoes carrying gene drives and antiparasitic effector molecules as tools for controlling pathogen transmission (1–4). These technologies must meet discovery, development, and delivery goals before acceptance as disease-control practices (5). Research with malaria vectors provides proofs-of-principle for this technology and its potential for real-world impact (1, 2, 4). However, work on development and delivery objectives is needed to define final products and performance. Target product profiles (TPP) have been drafted and provide the basis for evaluating products for field use (5, 6). TPPs comprise a list of key parameters, including drive efficiency, effector efficacy, genetic load (fitness), safety, production needs and release strategies, and a description of the ideal and the minimally-essential requirements for the product to have a practical value.
The gene-drive strain, AgNosCd-1, meets most essential TPP criteria for population modification of the African malaria vector, Anopheles gambiae. AgNosCd-1 has a drive efficiency as high as 100%, no major load, no apparent off-target effects, and a low frequency of generation of potential resistant alleles. Single releases of AgNosCd-1 males at ratios of 1:1 transgenic:wild-type (WT) achieved full introduction (every mosquito containing at least one copy of the gene-drive construct) in small cage laboratory trials within six generations, a period of ∼6 mo, well within a single annual malaria transmission cycle. AgNosCd-1 provides an excellent delivery vehicle that can be combined with antiparasite effector genes for development of strains for field trials.
Results
Generation and Molecular and Phenotypic Characterization of AgNosCd-1.
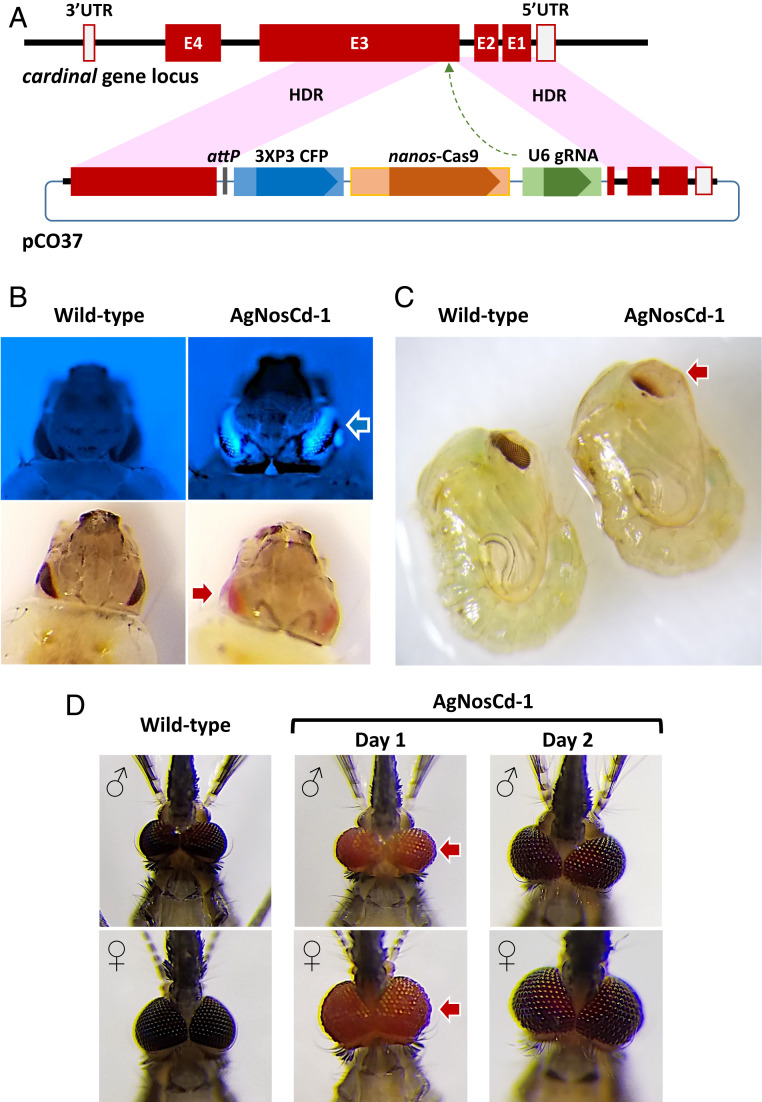
The gene drive plasmid, pCO37, targets the An. gambiae cardinal gene (Agcd), which encodes a heme peroxidase that catalyzes the last step in ommochrome biosynthesis, the conversion of 3-hydroxykynurenine to xanthommatin (7, 8) (Fig. 1 and SI Appendix, Fig. S1). pCO37 is designed to express Streptococcus pyogenes Cas9 (SpCas9) endonuclease using regulatory elements of the nanos gene ortholog from the An. gambiae PEST strain (9). The An. gambiae U6 gene promoter expresses a 23-nucleotide gRNA, 5′-GGTTAGCGACGATGCCAAGGCGG-3′, targeting Agcd exon 3. A 3XP3-CFP dominant marker cassette was used for identifying transgenic progeny. A φC31 attP sequence was included for subsequent recombinase-mediated modification of the integrated DNA. Agcd genomic DNA fragments (∼1 kilobase pairs [kb] in length) homologous to regions flanking the intended target cut site facilitate homology-directed repair (HDR)-mediated integration. Reagents and products are listed in SI Appendix, Tables S1 and S2, and procedures detailed in the SI Appendix, Materials and Methods.
Fig. 1.
Agcd gene, pCO37 gene-drive construct, and resulting phenotypes. (A) Agcd gene: maroon blocks, exons (E1-4); empty blocks, 3′- and 5′-untranslated regions (UTR); thick black line, introns and intergenic DNA. pCO37 plasmid: maroon blocks, homology arms from the Agcd gene; blue blocks, dominant marker gene components (3XP3 and CFP); tan blocks, drive components (nanos promoter and SpCas9 protein-encoding sequences); green blocks, guide RNA components (U6 promoter and gRNA sequence); dark gray block, φC31 attP “docking” site. Genes and features of pCO37 are not to scale and approximate sizes of components in kilobases are listed in the Materials and Methods. Recombination resulting from HDR initiated at the SpCaS9/gRNA-mediated cut site (broken-line arrow) occurring within the pink-shaded regions results in integration of the gene-drive construct. (B) CFP+ (blue arrow) and homozygous Agcd-mutant (red arrow) phenotypes in larvae and (C) Agcd-mutant (red arrow) phenotype in pupae. (D) Homozygous Agcd mutant phenotype “red eye” (red arrows) in adults. Approximate image magnifications for B, C, and D are 20, 10, and 20×, respectively.
pCO37 was coinjected with SpCas9 protein into 780 WT An. gambiae embryos. The 146 surviving G0 adults (18.7%) were outcrossed to WT members of the opposite sex and three transgenic G1 progeny (0.09%, 3 of 3,428) were positive (CFP+) for the marker gene. Two of these, one male and one female from a male founder family, survived. The insertion site was verified by sequencing the genomic DNA of the progeny of these founder mosquitoes, confirming that they had the expected SpCas9-mediated HDR integration that resulted in the insertion of ∼10 kb of exogenous DNA (SI Appendix, Fig. S2).
Agcd maps to the second chromosome (2R: 38742728–38749765) (https://www.vectorbase.org/) and mutations in it are recessive with homozygotes having a red-eye phenotype visible in larvae, pupae, and newly emerged adults (Fig. 1 B–D). A color gradient from posterior to anterior is seen in pupal eyes with the greatest accumulation of pigment close to the ocelli. The adult eye color darkens 1 d postemergence but does not achieve the deep-purple/black color of WT mosquitoes. AgNosCd-1 homozygotes showed changes during development in accumulation of ommochrome biosynthetic pathway metabolites with the most notable being a higher amount of xanthurenic acid in adults when compared to WT (SI Appendix, Fig. S1). The precursor, tryptophan, and intermediary metabolite, 3-hydroxykynurenine, also were increased in recently-emerged adults, pupae, and older adult AgNosCd-1, respectively, while kynurenic acid was higher in WT larvae.
AgNosCd-1 Drive Is Inherited Efficiently through Males and Females.
The CFP+ G1 male and female were pool-mated first with 20 WT females and the male then was mated separately with 20 WT females (Table 1). These two crosses show high HDR and drive efficiency, 95% (239 of 251) and 100% (50 of 50) introduction, respectively, in mosquitoes presumed to start as hemizygotes resulting from outcrosses. All subsequent intercrosses and outcrosses showed 100% introduction, except for 99.5% (1,656 of 1,663) in the G3 outcross. The progeny of these two crosses were identical and used to establish the AgNosCd-1 line.
Table 1.
Founder gene-drive experimental data
| Generation | Cross | Parents | Larvae | Pupae | ||||
| CFP+ | CFP− | CFP+ cd− | CFP+ cd+ | CFP− cd− | CFP− cd+ | |||
| G2 | OX/IX | G1 AgNosCd-1♂/G1 AgNosCd-1♀ 20WT ♀ | 239 | 12 | – | – | – | – |
| OX | G1 AgNosCd-1♂/20WT♀ | 50 | 0 | – | – | – | – | |
| G3 | IX | G2 AgNosCd-1♂/G2 AgNosCd-1♀ | 313 | 0 | 192 | 5 | 0 | 0 |
| OX | G2 AgNosCd-1♂/WT♀ | 151 | 0 | – | – | – | – | |
| G4 | IX | G3 AgNosCd-1♂/G3 AgNosCd-1♀ | 980 | 0 | 979 | 1 | 0 | 0 |
| OX | G3 AgNosCd-1♂/WT♀ | 1,656 | 7 | 0 | 181 | 0 | 0 | |
Except for the G4 intercross, pupae are only a sample of the larvae scored. No pupae from outcrosses were scored. Boldface columns and numbers are CFP+ mosquitoes. “–” indicates not examined. Abbreviations: cd−, cardinal (homozygous for mutant in gene giving a pale red-eye), cd+, cardinal (hemizygous for mutant in gene giving a WT eye color); CFP, cyan fluorescent protein (positive: +; negative: −); IX, intercrosses; OX, outcrosses.
Samples of pupae from the G2 and G3 intercrosses were scored for either WT or cardinal (cd, red) eye color, (Fig. 1B), and the percentages presumed to be homozygous for the AgNosCd-1 insertion (CFP+, red-eye color) were 97.5% (192 of 197) and 99.8% (979 of 980), respectively (Table 1). As expected from efficient drive and mutations in Agcd being recessive, a G3 outcross of AgNosCd-1 males to WT females produced 100% CFP+ WT-eye color mosquitoes (181 of 181). These data confirm that Agcd mutations are recessive and that homozygous disruptions of the locus produce an easily scored eye-color phenotype.
Forty male or female AgNosCd-1 hemizygotes were outcrossed separately to 40 opposite-sex WT to look for sex-specific lineage effects (SI Appendix, Fig. S3). The resulting male and female CFP+/WT-eye progeny were outcrossed serially to WT for three additional generations. Drive efficiencies varied from 85 to 100% with an average of 96.7% (8,367 of 8,629 of total progeny scored in F1-F4) (SI Appendix, Table S3). The frequency of mutant Agcd red-eye phenotypes should be zero as all of the matings were outcrosses. However, six progeny (6 of 8,629, 0.069%) had this phenotype, presumably resulting from somatic nonhomologous end-joining (NHEJ) mutations in the eye cells. These were less frequent than those reported in other drive systems in mosquitoes and Drosophila melanogaster (1, 2, 10). These data confirm high-efficiency drive and importantly, female founder lineages did not exhibit Mendelian segregation frequencies due to accumulation of resistance alleles in the germline, as seen previously (1).
These experiments revealed another eye phenotype, “tear,” and individuals from all crosses in which the gene-drive system came from males exhibited the phenotype at 0.29% (15 of 5,057), whereas it was 17-fold higher (4.9%, 174 of 3,572) in progeny from crosses with drive coming from females (SI Appendix, Fig. S4 and Table S3). This phenotype results from somatic mosaicism in the ommatidia and is not inherited through the germline (SI Appendix, Supporting Text and Tables S4–S6).
AgNosCd-1 Life-Time Parameters.
AgNosCd-1 mosquitoes did not exhibit a substantial genetic load compared to WT (SI Appendix, Table S7). WT and hemizygous or homozygous AgNosCd-1 did not show significant differences in the length of larval (∼2.7 d) and pupal (∼6 d) development counted from second-instar larvae (L2) onward, or larval viability (81 to 85%). Female AgNosCd-1 hemizygotes exhibited significantly higher fecundity (∼76 eggs per female; P < 0.05) than homozygous AgNosCd-1 (∼65 eggs per female) but not WT (∼72 eggs per female) and significantly higher fertility (∼71%; P < 0.001) than both WT (47% fertility) and homozygous AgNosCd-1 (52% fertility). No significant differences in fecundity or fertility were found between WT and homozygous AgNosCd-1 females (P > 0.05). Significant differences were observed in AgNosCd-1 males contributing to the next generation resulting from outcrosses (OX, >98% homozygous because of drive) and intercrosses (IX, homozygous) when competing with WT males for females in a one-generation trial (OX males: χ2 = 5.69, df = 1, n = 5,623, P < 0.05; IX males: χ2 = 143.58, df = 1, n = 7,023, P < 0.001). (“Contribution” as used here comprises aggregate factors, for example competitiveness and fertility, that influence the ability of male genetic material to be transmitted to the next generation.)
Long-Term Small Cage Trials of AgNosCd-1 Drive Dynamics.
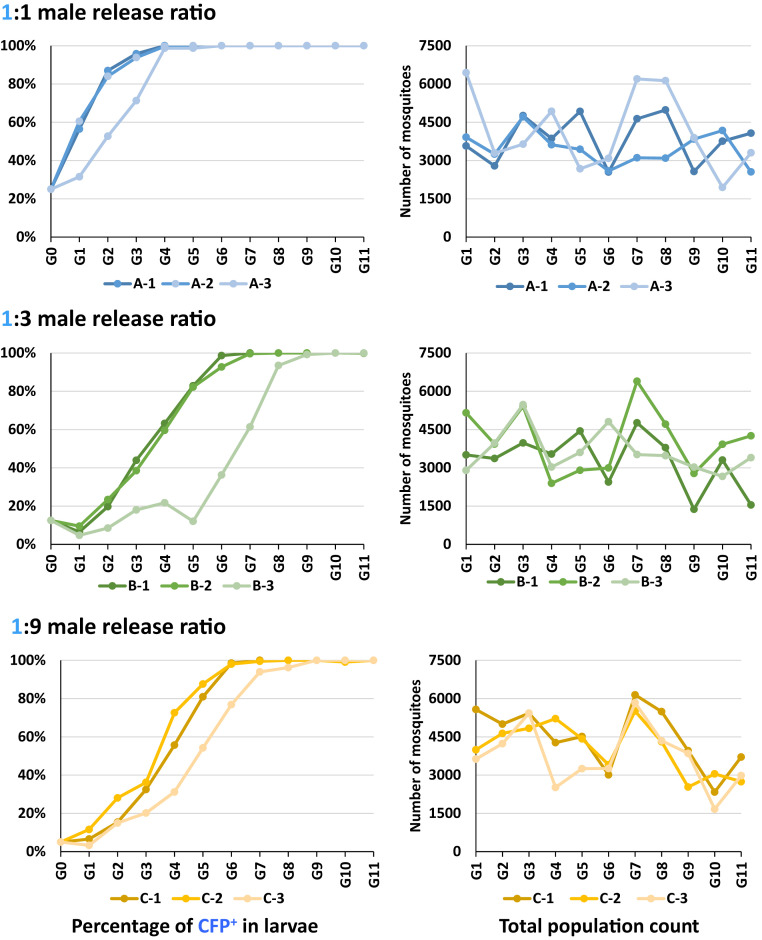
A series of nonoverlapping generation release trials were started by seeding triplicate cages with 150 WT adult females and 148 to 150 total males at ratios of 1:1 (A 1 to 3), 1:3 (B 1 to 3), and 1:9 (C 1 to 3) homozygous AgNosCd-1:WT (SI Appendix, Fig. S5). Three-hundred randomly-selected L2 from their progeny established each subsequent generation. An additional 600 L2 were reared and screened for CFP and eye-color phenotypes. A total of 384,872 mosquitoes were scored over 11 generations.
A 1:1 release ratio should produce ∼50% CFP+ progeny (one in two chance of a parental female mating with an AgNosCd-1 male) in the first-generation following release if there are equal contributions from parental AgNosCd-1 and WT males. We observed approximately equal, A-1, 56.40% (333 of 590; χ2 = 4.89 [P = 0.027]), or greater, A-2, 60.50% (305 of 504; χ2 =11.14 [P = 0.00084]) AgNosCd-1 contributions in two cages (Figs. 2 and 3 and SI Appendix, Tables S8–S10). Cage A-3 had a significantly lower AgNosCd-1 male contribution, 31.50% (175 of 556; χ2 = 38.16 [P < 0.00001]), a result consistent with those observed in the life-table hemi- and homozygous male contribution experiments (SI Appendix, Table S7).
Fig. 2.
Drive dynamics and population sizes of AgNosCd-1 mosquitoes in discrete, nonoverlapping generations based on small cage trials with 1:1, 1:3, and 1:9 gene-drive to WT males releases. Small cages (5,000 cm3) (A-1, A-2, A-3, B-1, B-2, B-3, C-1, C-2, and C-3) were seeded with 150 WT An. gambiae adult females and 148 to 150 AgNosCd-1 and WT males in ratios of 1:1, 1:3, and 1:9. The resulting next-generation progeny were scored as CFP+ (carrying the AgNosCd-1 gene drive cassette) or WT. Percentages of CFP+ mosquitoes (y axis) in the total population of each cage were scored at each generation (x axis) (Left). Total population size (y axis) in each replica cage at each generation (x axis) (Right).
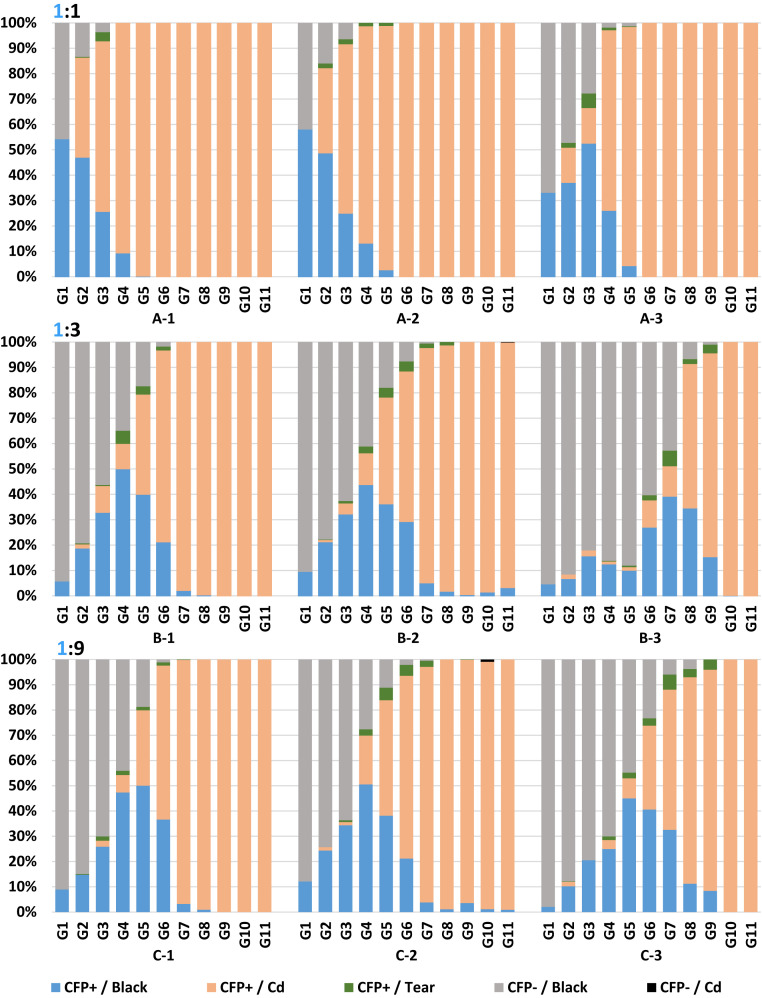
Fig. 3.
Distribution of mosquito phenotypes in small cage trials. Relative abundance in percentages (y axis) of eye color and CFP phenotypes of ∼600 randomly selected pupae per generation (x axis) in each cage (A-1, A-2, A-3, B-1, B-2, B-3, C-1, C-2, and C-3). Key: blue, CFP+ and WT eye color; salmon, CFP+ and cardinal Cd (red) eye color; green, CFP+ and tear (mosaic) phenotypes; gray, CFP− and WT eye color; black, CFP− and cardinal.
Cages A-1 and A-2 reached full introduction by G4 and cage A-3 at G6 (Figs. 2 and 3 and SI Appendix, Tables S8–S10). All lines were homozygous (CFP+/cd red-eyes) at G6. Overall population sizes fluctuated in the three cages with averages over 11 generations (excluding the G0) of 4,247, 3,829, and 4,553 in A1, A2, and A3, respectively. The average numbers of adult mosquitoes per generation (excluding the cage founders) surviving after the blood meal and potentially establishing the next generation were 247 (A-1), 237 (A-2), and 258 (A-3) with a combined average of 247 (82.3%).
First-generation CFP+ mosquitoes in the 1:3 cages were expected to be 25% (one in four chance of parental females mating with AgNosCd-1 males). All three cages had significantly lower AgNosCd-1 contributions: B-1, 6.54% (31 of 474; χ2 = 64.61 [P < 0.00001]), B-2, 9.50% (55 of 580; χ2 = 55.86 [P < 0.00001]), and B-3, 4.70% (24 of 508; χ2 = 83.53 [P < 0.00001]) (Figs. 2 and 3 and SI Appendix, Tables S11–S13).
A rapid increase in CFP+ mosquitoes followed immediately in the second-generation B-1 and B-2 cages with full introduction at G7 and G8, respectively. Cage B-3 is anomalous as CFP+ mosquitoes did not start to increase until G6, and it reached near full introduction by G10 (SI Appendix, Table S13). Homozygosity for the drive system was seen in G9 and G11 for cages B-1 and B-3, respectively. Cage B-2 had low numbers of CFP+/WT eye-color mosquitoes in G9 (0.5%, 3 of 589), G10 (1.5%, 7 of 466), and G11 (3.3%, 18 of 543). Population sizes averaged 3,605, 4,485, and 3,987 mosquitoes per generation (excluding cage founders) for B-1, B-2, and B-3, respectively (Fig. 2 and SI Appendix, Tables S11–S13). The average numbers of adult mosquitoes per generation potentially establishing the next generation were 236 (B-1), 254 (B-2), and 248 (B-3), with a total average of 246 (82%).
First-generation CFP+ percentages in the 1:9 release ratio cage population were expected to be 10% (1 in 10 chance of females mating with gene-drive males). Cages C1 and C3 had significantly lower AgNosCd-1 male contributions: 6.6% (370 of 5,574; χ2 = 63.00 [P < 0.00001]) and 3.3% (121 of 3,629; χ2 = 160.3 [P < 0.00001]), respectively (Figs. 2 and 3 and SI Appendix, Tables S14 and S16). Cage C-2 at 11.6% (462 of 3,992; χ2 = 9.88 [P = 0.0016]) had a slightly higher contribution (Figs. 2 and 3 and SI Appendix, Table S15). Remarkably, the gene-drive dynamics after the first generation were similar to the 1:3 release ratio. Full introduction was achieved by G7 to G9. Cages C-1 and C-3 were homozygous for the drive element by G9 and G10, respectively, and cage C-2 had not achieved this by G11, the end of the experiment. Fluctuating population sizes averaged 4,943, 4,465, and 4,101 per generation (founders excluded) for C-1, C-2, and C-3, respectively (Fig. 2 and SI Appendix, Tables S14–S16). The average numbers of adult mosquitoes potentially establishing the next generation were 258 (C-1), 254 (C-2), and 253 (C-3), with a total cage average of 255 (85%).
Exceptional phenotypes, no gene-drive element (CFP−), and red color (cd−) eyes, were recorded in cages B2 and C2 in G11 and G10, respectively (SI Appendix, Tables S12 and S15). Since previous cages in each series were near full introduction and nearly homozygous (97 to 98%), these mosquitoes could have homozygous or heteroallelic drive-resistant Agcd mutations. The single B2 G11 CFP−/cd− male (0.18%; 1 of 543) (SI Appendix, Table S12) was outcrossed to hemizygous drive (CFP+/cd+) females and the region adjacent to the gRNA target site sequenced for 12 of its CFP+/cd− progeny. Two distinct 11- or 14-bp deletions were identified indicating that the original male was heteroallelic (SI Appendix, Table S17). Because both include all or 2 of 3 nucleotides of the protospacer adjacent motif (PAM) site and the A residue 3 nucleotides to the 5′-end of the PAM site (−3 A), we expect these to be drive-resistant targets. Five CFP−/cd−, two males and three females (0.0091; 5 of 548), were recorded in C2 G10 larvae (SI Appendix, Table S15). Four of these were homozygous for one of two alternative deletion alleles of 11 (one mosquito) or 12 bp (three mosquitoes) (SI Appendix, Table S17). One deletion encompasses both the PAM site and the −3 A, and the other has an intact PAM site but is missing the –3 A. We expect both of these to be drive-resistant. Extrapolating a precise frequency for NHEJ formation of drive-resistant target site alleles in the germline from these data is not possible, but an estimate based on the homozygotes from the cage C2 G10 mosquitoes would be 0.095 (√0.0091) (SI Appendix, Table S15).
gRNA Target Variations in Mosquito Populations.
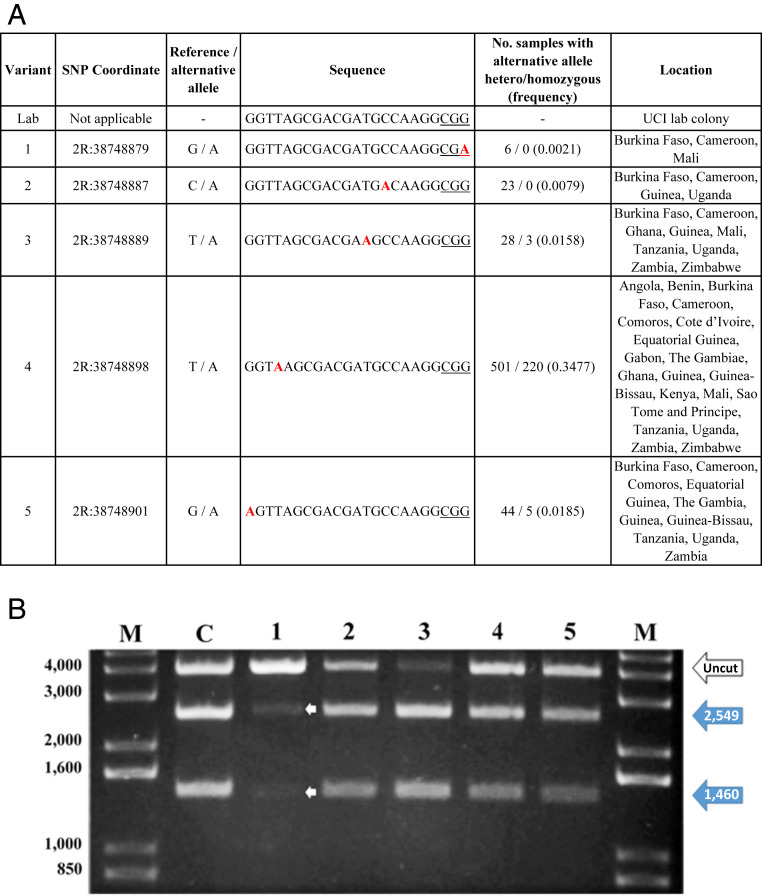
Twelve of the 23 positions in the gRNA target site showed nucleotide variation in colonized and field-derived specimens of An. gambiae and Anopheles coluzzii (SI Appendix, Table S18). Two sites, one each in the PAM (2R:38748880) and in the seed region (2R:38748883) had three alternate alleles. Sites 2R:38748880, 2R:38748883, and 2R:38748885 were polymorphic in single individuals in one of the An. gambiae or An. coluzzii samples. One half of the polymorphic sites (2R:38748881, 2R:38748883, 2R:38748887, 2R:38748889, 2R:38748898, 2R:38748901) had >1% alternative alleles, with 2R:38748898 at 43% being the most abundant in one sample from the Ag1000G project. However, only one site, 2R:38748883, had an SNP >1% in a potentially high-impact region and this was only in the highly-inbred G3 laboratory colony line. Furthermore, screening of the entire Agcd exon 3 failed to find a target site without any polymorphisms that would fit canonical target needs (moderate GC-content, no off-targets), underlining the relatively high quality of the target site chosen (11).
Four sites (2R:38748987, 2R:38748989, 2R:38748998, and 2R:38748901) were selected for SpCas9/gRNA-mediated cleavage in vitro based on their frequency (>1%) in samples and one, 2R:38748979, for being in the predicted high-impact PAM region (12). 2R:38748981 was not included because SpCas9 cleaves target sites with variants in the third nucleotide of the PAM sequence (12, 13). All sites were cleaved, resulting in 2,549 and 1,460 bp fragments indicative of SpCas9/gRNA activity although the PAM site variant appeared qualitatively to be cleaved less efficiently (Fig. 4). The geographical distribution of the tested nucleotide polymorphisms included 26 countries throughout the African continent.
Fig. 4.
Agcd SNP analysis in wild mosquito populations derived from Africa and SpCas9/gRNA-directed cleavage analysis in vitro. (A) Chromosome coordinates, nucleotide change from the reference allele, nucleotide sequence (5′-3′ orientation) with SNPs in bold red, number of samples heterozygous/homozygous for the SNP, and SNP frequency and sample source. The laboratory variant is from the WT G3 colony. (B) In vitro cleavage assay testing the efficiency of SpCas9/gRNA cleavage of target site SNP variants found in populations from Africa. Plasmids, pCO-57, pCO-58, pCO-59, pCO-60, pCO-61, and pCO-62, containing control (C) WT laboratory colony and variant gRNA sites 1 to 5, respectively, were linearized and used in reactions with SpCas9 and the gRNA. Products at 2,549 and 1,460 bp (large solid blue arrows) are indicative of SpCasp9/gRNA-directed cleavage (uncleaved plasmids are indicated by the large white arrow). Small white arrows show cleavage products in variant 1. M, molecular weight markers.
Off-Target Effects in AgNosCd-1.
Three potential off-target sites, all on the second chromosome, were predicted in the PEST genome sequence (https://www.vectorbase.org/): off-1 is in AGAP004761 (predicted cut site: 2L 3,130,338), off-2 is in the intergenic region between AGAP002460 and AGAP002461 (predicted cut site: 2R 21,710,305), and off-3 is in AGAP003578 (predicted cut site: 2R 40,155,032) (SI Appendix, Fig. S6). All sites have a thymine (T) for cytosine (C) transition in the third positions of the PAM site when compared to the canonical Agcd target sequence. off-1 and off-3 have four additional nucleotide differences, none of which are shared in common. Consistent with its intergenic location, off-2 was polymorphic in the WT colony with two alternatives, off 2-1, off 2-2, sharing four of five total nucleotide differences, and the third, off 2-3, with six, three of which were common to all off-2 variants. Only off-target 1 and the positive control were cleaved in the SpCas9/gRNA complex in vitro assay (SI Appendix, Fig. S6). No indels related to any of the predicted off-target sites were found in vivo in 25 AgNosCd-1 samples (G8: 5 males and 20 females) randomly selected from the cage trials using gene amplification with specific primers (off-1_F1 and R1, off-2_F1 and R1, and off-3_F1 and R) and Sanger sequencing (SI Appendix, Table S2 and Fig. S7).
Further testing in vivo of off-1 used next-generation sequencing and a larger sample set of AgNosCd-1 individuals from the cage trials (52 G9 [20 males/32 females] and 176 G10 [84 males/92 females]) (SI Appendix, Fig. S8). Low deletion frequencies were detected in control WT (0.06%; 33 1-bp deletions, total 33 of 54,447 reads), G9 females (0.04%; 20 1-bp and 1 2-bp deletions, total 21 of 47,887 reads), G9 males (0.03%; 16 1-bp and one each of 2-, 4-, and 5-bp deletions, total 19 of 54,708 reads), G10 females (0.05%; 28 1-bp and one each of 2- and 4-bp deletions, total 30 of 56,861 reads), and G10 males (0.05%; 18 1-bp and 1 4-bp deletions, total 19 of 37,800 reads). Not all of the observed deletions can be attributed to SpCas9 activity as some occurred in the WT samples that were not exposed to the nuclease–gRNA complex. Considering only deletions at or within 3 nucleotides around the Cas9 cut-site and not present in the WT samples, only four reads qualified as deletions that could be due to Cas9 cleavage and subsequent NHEJ repair: two different single-read events in G9 males (0.0036% of total reads; 2 of 54,708), a single-read event in G10 females (0.0017% of total reads; 1 of 56,861), and a single-read event in G10 males (0.0026% of total reads; 1 of 37,800) (SI Appendix, Fig. S9). Thus, the total overall off-target deletions at this site are ∼0.0027% (4 of 149,369).
Discussion
AgNosCd-1 meets key TPP parameters for gene-drive system components that include the physical elements of the drive system (Table 2). Specifically, the sources of the DNA, and their size and complexity minimize foreign DNA, while keeping the element as small and simple as possible. All DNA sequences are from An. gambiae, except for the SpCas9 ORF, fluorescent marker cassette, site-specific recombination docking sequence, transcription terminator sequences, and a small amount of plasmid DNA in spacer regions. The overall size, ∼10 kb, of the integrated DNA is within limits of other systems that show robust drive (1, 14), and the complexity is minimal, with five drive components and two more for future effector molecules not included in this study. The drive components were stable during prolonged small cage trials with no failure due to recombination or mutation. However, this will have to be monitored continuously as the product is scaled up to include the effector gene sequences.
Table 2.
Summary of some gene-drive system parameters with ideal, minimally essential and AgNosCd-1 performance features
| Parameter | Ideal | Minimally Essential | AgNosCd-1 |
| Source DNA | All orthologous from target species | All control DNA functional and stable | All DNA from An. gambiae except the SpCas9 ORF, φC31 site-specific recombination (attP) sequence, 3xP3marker gene-CFP, SV40 transcription termination sequences. Spacer DNA may originate in bacterial plasmids or be synthetic. |
| Size | Small as possible for efficacy | Same as ideal | Cargoes (gene-drive components and marker genes) are ∼10 kb in length, similar in size to previously successful constructs (1, 14). |
| Complexity | Fewest components as possible to assure efficacy and safety | Same as ideal | The gene drive construct has five components, nosCas9, U6gRNA, 3XP3, CFP, lox site, and SV40. |
| Stability | No breakdown due to mutation or recombination | Rate of breakdown does not preclude use in local elimination | No evidence of breakdown in outcrosses, intercrosses and small cage trials. |
| Drive target locus | Highly conserved sequence in gene critical for parasite development, a mutation in which imposes no genetic load on the mosquito | Conserved and no major load | cardinal gene ortholog, Agcd, is conserved in colonized and wild mosquitoes, and has no gross load. |
| Male competitiveness | Equal or more than WT | >20% of WT | Heterozygous AgNosCd-1 males may outperform WT and transgenic homozygous males. |
| Female fecundity | Equal or more than WT | >20% of WT | Heterozygous AgNosCd-1 females outperformed homozygotes and WT controls in both fecundity and fertility. |
| Strain maintenance | Maintained as homozygous strain | Balanced heterozygous strain or outcross at every generation | AgNosCd-1 can be maintained as homozygous. |
| Drive inheritance | Both sexes | One | AgNosCd-1 is active in both males and females |
| Efficacy (percent population carrying gene) | 100% full introduction into target wild populations | ≥90% | 85–99% through female lineages, |
| 97–100% through male lineages | |||
| Time to full introduction | <1 y | 2–8 y (26) | 1:1 introductions of AgNosCD-1 males to WT result in full introduction within six generations, ∼6 mo. |
| Impact of resistant drive targets | No resistant targets | No impact on effector efficacy | SNP variants in colonized and wild-derived mosquitoes cleaved by SpCas9/gRNA in vitro. No effects seen in small cage trials. |
| Off-target drive effects | No off-target effects | To be determined by target and load properties | Low frequency of NHEJ-induced indels at only one of three off-target sites. |
| Nontarget drive effects | No nontarget effects | Nonfunctional in other species | No sequence identities in the gRNA targeting site in mosquitoes other than the An. gambiae complex. |
Parameters adapted from Carballar-Lejarazú and James (5).
Other key parameters that were fulfilled included the gRNA target site, genetic load, and possible off-target and nontarget effects. The ideal target site, highly conserved, in a gene whose product is critical for malaria parasite development and one in which a mutation would not confer a load, is not likely to exist. Highly-conserved genes are likely to be under negative selection and their disruption could impose a load. We expected and observed a recessive phenotype from mutations at the AgNosCd-1 gRNA target, which could be useful for tracking gene drive in the field. While there were effects on accumulation of ommochrome pathway metabolites in homozygous mutant Agcd mosquitoes, the resulting red-eye phenotype darkens to near WT in older adults. This is likely due to an alternate enzyme being able to carry out the function of the heme oxygenase in these mosquitoes. Viability and length of time of the subadult stages and adult longevity were not significantly different among AgNosCd-1 homozygote, hemizygote, and WT mosquitoes. Importantly, while there were some effects on male contribution and female fecundity, no aggregate load impacted overall drive dynamics in the small cage trials. Furthermore, two of the three cage trials with 1:9 gene-drive to WT males showed full introduction within 10 generations. If all gene-drive males were equally as competitive as WT, this would mean they had a 10% chance of contributing to the next generation. Therefore, we might expect that at release ratios of 1:1 (gene-drive to WT), future genetic loads imposed by adding effector genes that result in aggregate fitness reductions to 20% of WT would still allow full introduction. Thus, the inheritance bias of drive systems mitigates the need for excessive numbers of release animals to offset fitness costs. The negligible loads evidenced here also enable the strain to be kept as homozygotes, greatly reducing the effort required to rear and maintain the transgenic line.
AgNosCd-1 drives efficiently in both adult males and females, with males narrowly out-performing females most likely from the action of SpCas9/gRNA complexes on incoming male chromosomes in embryos from drive-positive females producing NHEJ mutations (1, 14). This phenomenon also results in the production of somatic mosaic mosquitoes evidenced here by the tear phenotype.
nos regulatory elements provide more precise germline-restricted expression than those from vasa and mitigate high-frequency maternal effects and NHEJ drive-resistant alleles, as observed previously (1, 9). Previous work in mosquitoes showed that nos genes were expressed preferentially in females with mRNA levels undetectable in males by Northern blot analyses and some gene-amplification protocols (15, 16). However, Northern blot, microarray, and hybridization in situ analyses detected mRNA in adult males of other Diptera (17–19), and it is clear from the results here that there is expression in An. gambiae males. Male nos expression also was observed in transgenic Anopheles stephensi using a different configuration of the nos control DNA to drive the expression of transposases (20).
Times to full introduction, as short as four generations with the 1:1 AgNosCd-1:WT male release ratio, are good and this could be improved further in field trials by dispersed, multiple releases. Studies of mosquito abundance at prospective trial sites will provide data on the size of target wild populations and inform this protocol. Previous studies on abundance and densities of mosquitoes in the wild vary depending on the physical unit (for example, village, district, or other) and the time of year. It has been possible with other technologies (Wolbachia-enhanced strains and sterile insect techniques) to rear large quantities of release mosquitoes, and we anticipate that we can achieve the numbers needed to have an impact within one malaria transmission season (21–25). Modeling supports the conclusion that even with a protracted period of up to 8 y, there would still be an epidemiologically significant impact on malaria transmission (26).
The impact of drive-resistant gRNA target site variants on the efficiency of Cas9/gRNA-based gene-drive systems is debated vigorously (1, 2, 4, 12, 26–28). Variants may either be preexisting in wild populations or generated by NHEJ outcomes during drive. The An. gambiae genome is highly polymorphic in natural populations (11, 29), and therefore potentially prone to the former challenge. However, screening of the gRNA target site in hundreds of mosquitoes from natural populations throughout Africa revealed a high level of sequence conservation with most PAM and adjacent nucleotides with alternative allele frequencies <1%, and most of these tend to be confined to one to three countries. Furthermore, five of the most abundant or predicted to be critical for drive were cleavable in vitro by SpCas9/gRNA complexes. While the extent of cleavage cannot be related to the efficiency of drive in vivo, the fact that they can be cut supports a hypothesis that these variants would only slow the drive and not prevent AgNosCd-1 from spreading an effector gene through a target population. No resistance alleles with an apparent selective advantage emerged to affect drive during cage trials, an observation consistent with the negligible loads associated with disruptions of the target Agcd locus. These combined observations support the conclusion that naturally-prevalent or NHEJ-induced resistance alleles are not likely to prevent using AgNosCd-1 to deliver an effector gene cargo. However, field trials utilizing naturally-confined populations need to be conducted to test this prediction (11).
The impact of AgNosCd-1 off-target effects appears minimal in the experiments we have conducted so far. Only one of five off-targets most similar to the canonical site was cleavable in vitro by SpCas9/gRNA complexes. Sequencing a large number of samples containing this variant showed a low frequency of indels that could be associated with off-target cleavage. These indels did not increase in frequency, and therefore were not selected during the cage trials. The functions of the two genes with the putative off-target sites are not known or described, and therefore any phenotypes that would manifest from their disruption also are unknown. Interestingly, the predicted intergenic off-target 2 site showed the highest variability as might be expected in a region not under negative selection.
Nontarget effects have been raised by other investigators as a potential concern (30). While unreported BLAST and other analyses show no sequence identities with the Agcd targeting site in mosquitoes other than the An. gambiae complex, sequences identical to 17 bp of the cd target site including the PAM are found in Drosophila species and salmonids. Other highly similar sequences (up to 20 bp including PAM) are found in bacteria, fungi, and plants. It is not expected that An. gambiae nos 5′-end control DNA will function properly in any of these organisms as the gene structure is highly divergent, and WT orthologs of nos genes from Dipteran species, including the closely-related Drosophila virilis, failed to fully complement a null phenotype in transgenic D. melanogaster (31). We will look at this empirically by attempted reciprocal mating of AgNosCd-1 with colonized An. gambiae strains (for introduction) and closely-related species (for introgression). Our prediction is that AgNosCd-1 can be introduced into the An. gambiae strain variants and perhaps introgressed into those species that can form fertile hybrids [An. coluzzii and Anopheles arabiensis (32, 33)]. However, fertile hybrids are a prerequisite to introgression, so any species showing this reproductive barrier should be immune to the drive.
In summary, the data here show that AgNosCd-1 meets tested gene-drive criteria for a TPP and support the further development of it as a system for a population modification trial. Additional criteria consistent with a phased testing of the potential product (34–36) include defining effector molecule efficacy, additional safety features (e.g., nonmalaria pathogen transmission), entomological and epidemiological endpoints for field testing, field-trial modeling parameters and outcomes, and strain utilization and production requirements. The processes and outcomes of evaluating these criteria with potential end-users and stakeholders will inform concomitant efforts in hazard identification and risk assessment, regulatory appraisal, and community engagement.
Materials and Methods
Detailed procedures are listed in SI Appendix, Materials and Methods.
An. Gambiae Cardinal Gene (Agcd) Target Site Selection.
The An. gambiae heme peroxidase 6 gene (AgHPX6, AGAP003502) is the ortholog of the cardinal (cd) locus in D. melanogaster and was chosen as the site for gRNA-directed insertion of the gene-drive system. Agcd genomic sequences from the An. gambiae PEST (AgamP4) strain were analyzed using specific oligonucleotide primers for potential gRNA target and associated off-target sites.
Plasmid Design and Construction.
Standard molecular biological technologies were used for the isolation and cloning of specific DNA fragments to construct the gene drive system. Sizes in base pairs of specific fragments for pCO-37 are Agcd homology arm 1 (HM1) derived from exon 3, 1,067 bp; 3xP3 promoter, 265 bp; CFP, 720 bp; SV40 polyA, 122 bp; An. gambiae nos promoter/5′-end UTR, 1,642 bp; SpCas9 ORF, 4,104 bp; An. gambiae nos 3′-end UTR, 642 bp; An. gambiae U6 promoter/5′-end UTR, 600 bp; gRNA, 20 bp; gRNA scaffold, 76 bp; An. gambiae U6 3′-end UTR, 378 bp and Agcd HM2 spanning exon 1 to the 5-end of exon 3 and intervening introns, 1,167 bp (Fig. 1).
Microinjection of Embryos and Screening Procedures.
Mosquitoes were injected with a solution containing 300 ng/μL of pCO-37, and 100 ng/μL of SpCas9 protein (PNA BIO Inc). Resulting adult G0 males and females were outcrossed to WT mosquitoes of the opposite sex in pools of ∼5 G0 males or 15 to 30 G0 females and progeny screened for CFP fluorescence using UV-fluorescence microscopy.
Molecular Validation of Gene-Drive Cassette Integration.
Validation of precise integration was confirmed by amplification analysis using specific oligonucleotide primers. Mosquitoes hemizygous (harboring one copy of the dominant CFP fluorescence gene) cannot be readily distinguished from homozygotes (harboring two copies); therefore, left and right portions of the homology arm sequences were amplified to ascertain the presence of a WT Agcd allele.
Male and Female Lineage Gene-Drive Dynamics.
The roles of maternal effects and targeted SpCas9 activity at the Agcd gene in somatic cells were assessed by crossing males and females hemizygous (originating from an outcross) for the AgNosCd-1 cassette with WT mosquitoes of the opposite sex and scoring the progeny for eye phenotype, including the presence/absence of the CFP marker gene. Progeny with WT color eyes (deep purple/black) and the CFP marker gene (CFP+) were recovered as virgin males and females and crossed again to WT mosquitoes. Male and female founder lineages were maintained until the fourth generation of progeny had been screened or two successive generations of males had produced progeny.
Mass Spectrometry.
Metabolite quantification in the ommochrome biosynthetic pathway was carried out at the University of California, Irvine, Mass Spectrometry Facility on a Waters Aquity UPLC system and Waters Quattro Premier XE mass spectrometer using an Aquity UPLC CSP C18 1.7-μM column and guard column. Metabolite extraction was performed by submerging 10 adult mosquitoes (WT and AgNosCd-1 males and females) in 500 μL of methanol with 1% formic acid (vol/vol) at 25 °C for 24 h.
Tear Intercrosses.
All mosaic phenotype progeny (tear) from G3 (11 males, 31 females), G4 (18 males, 29 females), and G5 (11 males, 38 females) of the AgNosCd-1 cage trials, in addition to 14 males and 16 females of the sex specific-effect experiments, were intercrossed, with each separate generation representing an experimental replicate. Mosaic phenotype mosquitoes were identified during pupation as part of the screening protocol for the cage trials and general screening for the maternal-effect experiment.
Digital Droplet PCR Drop-off Assay.
Digital droplet PCR drop-off assay reactions were performed as described in Carballar-Lejarazú et al. (37).
Life Table Parameters.
Life table parameters were assessed in AgNosCd-1 (hemizygous [AgNosCd-1-OX] and homozygous [AgNosCd-1-IX]) mosquitoes and compared with the WT strain. Each experiment was performed in triplicate unless specified otherwise.
Cage Trials.
Triplicate (1 to 3) 5,000-cm3 cages were seeded with three single-release ratios of homozygous AgNosCd-1-IX to WT adult males: A series: 1:1 (75:75) (AgNosCd-1-IX:WT), B series: 1:3 (37:111), and C series: 1:9 (15:135), and WT adult females added to each cage to achieve an equal male:female ratio with a total population of ∼300 individuals per cage. Subsequent screening and analysis were performed as described in Pham et al. (4).
Variability of Agcd Target Site.
Whole-genome sequencing data of individual mosquito were generated for 120 An. gambiae sensu stricto samples from natural populations originating from Mali, Cameroon, Tanzania, Zambia, and the Union of Comoros. Additionally, 100 samples from the closely related species, An. coluzzii, from natural populations found in Mali, Benin, Equatorial Guinea, Cameroon, and São Tomé and Príncipe were screened. All genomic DNAs were sequenced on an Illumina HiSEq. 4000 instrument. SNPs were called with Freebayes v1.2.0 (38). The publicly-available polymorphism data from the Anopheles gambiae 1000 Genomes Consortium (AG1000G,2 AR1 data release) comprising calls from whole-genome sequence data from 590 An. gambiae specimens from Burkina Faso, Cameroon, Gabon, Guinea, Kenya, and Uganda was obtained. Additional An. coluzzii specimens from Angola and Burkina Faso were included.
SpCas9 In Vitro Cleavage Assay of WT and SNP Variant Target Sites.
An in vitro assay tested the ability of the gRNA/SpCas9 complex to cleave the target sites containing SNPs found in wild populations of African An. gambiae. Forward and reverse oligonucleotide pairs for each DNA target sequence version were annealed and used to amplify double-stranded DNA fragments that then were cloned into the pCR4-TOPO vector (Invitrogen) via the TA cloning strategy. The target plasmids were linearized with NcoI and mixed with 300 ng of SpCas9/cardinal-gRNA mix and the resulting products resolved on agarose gels.
Off-Target Site Analyses.
Analysis in silico of DNA sequences and design of primers were performed using the Snapgene (https://www.snapgene.com:443/) bioinformatics software. Plasmids with putative off-target sites were used for the SpCas9 cleavage assay in vitro, as described in the assay of SpCas9 In Vitro Cleavage Assay of WT and SNP Variant Target Sites.
Sequencing Mosquito Samples.
Sanger and next-generation sequencing were used to analyze potential off-target cleavages in genomic DNA from mosquitoes recovered from the cage trials.
Supplementary Material
Acknowledgments
We thank Judy Coleman, Kristy Hwang, Drusilla Stillinger, Kiona Parker, Sean Firth, and Laura Partida for mosquito husbandry; Hans Gripkey, Travis Collier, Abram Estrada, and Melina Campos for the analysis of target-site polymorphisms; and Rhodell Valdez for help with the manuscript. Funding was provided by the University of California, Irvine Malaria Initiative and the Bill and Melinda Gates Foundation (OPP1160739). A.A.J. is a Donald Bren Professor at the University of California, Irvine.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010214117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Gantz V. M.et al., Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. U.S.A. 112, E6736–E6743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond A.et al., A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyrou K.et al., A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pham T. B.et al., Experimental population modification of the malaria vector mosquito, Anopheles stephensi. PLoS Genet. 15, e1008440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carballar-Lejarazú R., James A. A., Population modification of Anopheline species to control malaria transmission. Pathog. Glob. Health 111, 424–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James S. L., Marshall J. M., Christophides G. K., Okumu F. O., Nolan T., Toward the definition of efficacy and safety criteria for advancing gene drive-modified mosquitoes to field testing. Vector Borne Zoonotic Dis. 20, 237–251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Beerntsen B. T., James A. A., Oxidation of 3-hydroxykynurenine to produce xanthommatin for eye pigmentation: A major branch pathway of tryptophan catabolism during pupal development in the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 29, 329–338 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Han Q., Beerntsen B. T., Li J., The tryptophan oxidation pathway in mosquitoes with emphasis on xanthurenic acid biosynthesis. J. Insect Physiol. 53, 254–263 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meredith J. M., Underhill A., McArthur C. C., Eggleston P., Next-generation site-directed transgenesis in the malaria vector mosquito Anopheles gambiae: Self-docking strains expressing germline-specific phiC31 integrase. PLoS One 8, e59264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champer J.et al., Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 13, e1006796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt H.et al., Abundance of conserved CRISPR-Cas9 target sites within the highly polymorphic genomes of Anopheles and Aedes mosquitoes. Nat. Comm. 11, 1425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drury D. W., Dapper A. L., Siniard D. J., Zentner G. E., Wade M. J., CRISPR/Cas9 gene drives in genetically variable and nonrandomly mating wild populations. Sci. Adv. 3, e1601910 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karvelis T.et al., Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 16, 253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adolfi A.et al., Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. 10.1101/2020.08.02.233056 (3 August 2020). [DOI] [PMC free article] [PubMed]
- 15.Calvo E.et al., Nanos (nos) genes of the vector mosquitoes, Anopheles gambiae, Anopheles stephensi and Aedes aegypti. Insect Biochem. Mol. Biol. 35, 789–798 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Juhn J., Marinotti O., Calvo E., James A. A., Gene structure and expression of nanos (nos) and oskar (osk) orthologues of the vector mosquito, Culex quinquefasciatus. Insect Mol. Biol. 17, 545–552 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C., Lehmann R., Nanos is the localized posterior determinant in Drosophila. Cell 66, 637–647 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Guerrero F. D.et al., Microarray analysis of female- and larval-specific gene expression in the horn fly (Diptera: Muscidae). J. Med. Entomol. 46, 257–270 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Chau J., Kulnane L. S., Salz H. K., Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc. Natl. Acad. Sci. U.S.A. 109, 9465–9470 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macias V. M.et al., nanos-Driven expression of piggyBac transposase induces mobilization of a synthetic autonomous transposon in the malaria vector mosquito, Anopheles stephensi. Insect Biochem. Mol. Biol. 87, 81–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho D. O.et al., Suppression of a field population of Aedes aegypti in Brazil by sustained release of transgenic male mosquitoes. PLoS Negl Trop. Dis. 9, e0003864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan P. A.et al., Establishment of wMel Wolbachia in Aedes aegypti mosquitoes and reduction of local dengue transmission in Cairns and surrounding locations in northern Queensland, Australia. Gates Open Res. 3, 1547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benedict M. Q.et al., Pragmatic selection of larval mosquito diets for insectary rearing of Anopheles gambiae and Aedes aegypti. PLoS One 15, e0221838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benedict M. Q.et al., Colonisation and mass rearing: Learning from others. Malar J. 8, S4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford J. E.et al., Efficient production of male Wolbachia-infected Aedes aegypti mosquitoes enables large-scale suppression of wild populations. Nat. Biotechnol. 38, 482–492 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Eckhoff P. A., Wenger E. A., Godfray H. C., Burt A., Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. Proc. Natl. Acad. Sci. U.S.A. 114, E255–E264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callaway E., Gene drives meet the resistance. Nature 542, 15 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Unckless R. L., Clark A. G., Messer P. W., Evolution of resistance against CRISPR/Cas9 gene drive. Genetics 205, 827–841 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Anopheles gambiae 1000 Genomes Consortium, Ag1000G phase 2 AR1 data release. http://www.malariagen.net/data/ag1000g-phase2-ar1. (2017). Accessed 21 March 2019.
- 30.Roberts A.et al., Results from the workshop “Problem formulation for the use of gene drive in mosquitoes. Am. J. Trop. Med. Hyg. 96, 530–533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtis D., Apfeld J., Lehmann R., Nanos is an evolutionarily conserved organizer of anterior-posterior polarity. Development 121, 1899–1910 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Hanemaaijer M. J.et al., Introgression between Anopheles gambiae and Anopheles coluzzii in Burkina Faso and its associations with kdr resistance and Plasmodium infection. Malar J. 18, 127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernardini F., Kriezis A., Galizi R., Nolan T., Crisanti A., Introgression of a synthetic sex ratio distortion system from Anopheles gambiae into Anopheles arabiensis. Sci. Rep. 9, 5158 (2019). Erratum in: Sci. Rep.9, 7915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization , Guidance Framework for Testing of Genetically Modified Mosquitoes, (WHO/TDR Publications, 2014). [Google Scholar]
- 35.Akbari O. S.et al., BIOSAFETY. Safeguarding gene drive experiments in the laboratory. Science 349, 927–929 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Academies of Sciences, Engineering, and Medicine , Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values, (The National Academies Press, Washington, DC, 2016). [PubMed] [Google Scholar]
- 37.Carballar-Lejarazú R., Kelsey A., Pham T. B., Bennett E. P., James A. A., Digital droplet PCR and IDAA for the detection of CRISPR indel edits in the malaria species Anopheles stephensi. Biotechniques 68, 172–179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrison E., Marth G., Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 (20 July 2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.