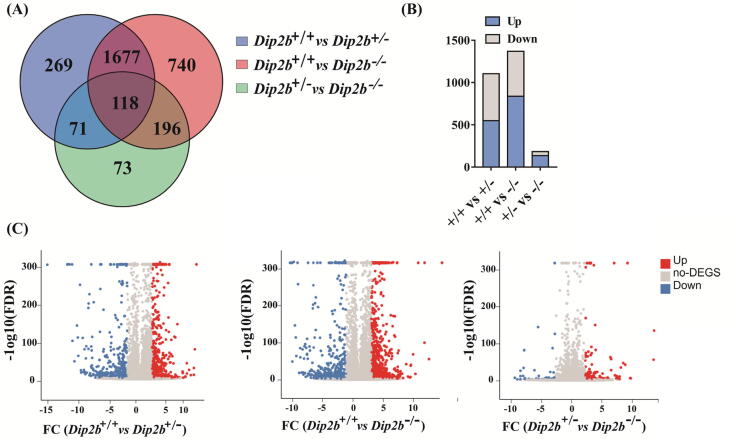
Fig. 2.
Overview of gene expression profiling. (A) Venn diagram showing unique and overlapping DEGs between heterozygous and homozygous MELFs (FC ≥ 1 and FDR ≤ 0.001). (B) Number of up- and down-regulated DEGs in Dip2b−/− and Dip2b+/− vs to Dip2b+/+ (C) Volcano plots highlighting significant DEG among three comparative samples. Each dot in plot corresponds to one differentially expressed gene, the y-axis represents −log10 (FDR) and the x-axis displays the differences of FC values in samples. Blue and red dots represent up- and down-regulated differentially expressed genes, whereas gray dots indicate genes with no change. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)