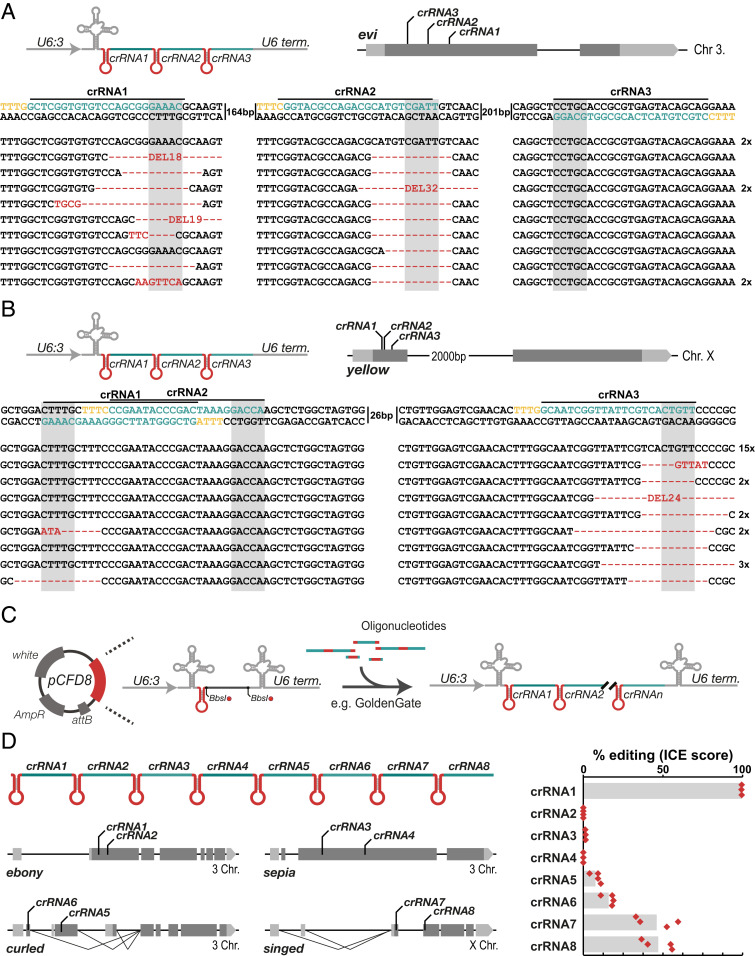
Fig. 2.
Multiplexed gene editing through crRNA arrays. (A) Gene editing of evi with three arrayed crRNAs. crRNAs were expressed from a U6:3 promoter and combined with nos-Gal4 UAS-LbCas12a transgenes for germline-restricted mutagenesis. Flies were raised at 29 °C and viable, indicative of efficient restriction of gene disruption to the germline. nos-Gal4 UAS-LbCas12a pCFD8-evi3x males were crossed to wild-type virgin females and genomic DNA from individual offspring was subjected to PCR amplification of the evi locus. PCR amplicons were sequenced and sequencing traces were parsed into wildtype and alternative allele sequences (Materials and Methods). The majority of animals harbor mutant evi alleles modified at the target sites of crRNA1 and crRNA2. Edited sequences are indicated in red and the crRNA target site is indicated in green, the PAM in yellow, and the window in which LbCas12a is expected to cut is shaded in gray. Indels at individual target sites are typically larger than 10 bp, further supporting data presented in SI Appendix, Fig. S4. (B) Gene editing of y with three arrayed crRNAs. The experiment was conducted as described above using a pCFD8-y3x transgene. Sequencing of PCR amplicons revealed highly efficient gene editing at the target site of the third crRNA in the array and editing with low efficiency at the target site of the first crRNA. No mutations were detected at the target site of the second crRNA. (C) Due to the compact size of Cas12a, several crRNAs can be encoded on commercially available oligonucleotides and oligos can be fused to construct larger crRNA arrays encoded in pCFD8 (see SI Appendix, Supplementary Cloning Protocol). (D) Multiplex gene targeting of four genes with two crRNAs each. Flies transgenic for a 8x crRNA array and act5C-LbCas12a were raised at 29 °C and had ebony cuticle and bristles displaying the singed phenotype. Each target locus was PCR amplified and amplicons were subjected to Sanger sequencing. Efficiency of gene editing was inferred from sequencing traces by ICE analysis (Materials and Methods). High levels of activity were detected for crRNA1 targeting e, crRNAs 7 and 8 had intermediate activity and crRNAs 5 and 6 showed low to medium activity. crRNAs 2 to 4 were inactive. crRNAs 1 (crRNA-e in Fig. 1 C and D), 2 (crRNA-e6) and 4 (crRNA-se) were in parallel also tested when expressed as single crRNAs in pCFD8 (Fig. 1D) with consistent results.