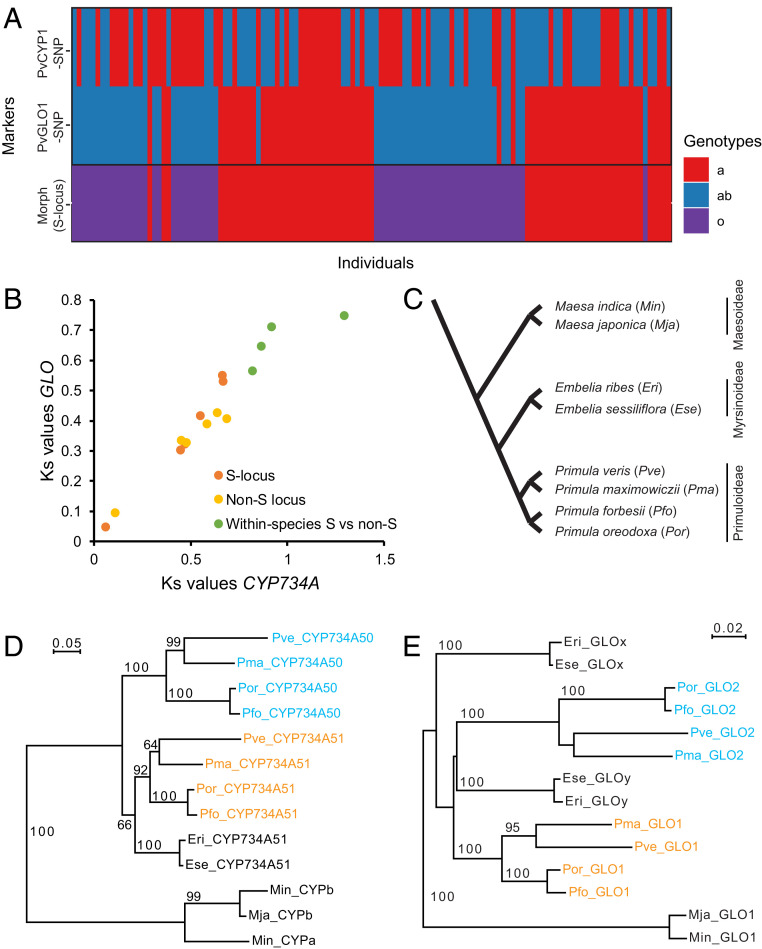
Fig. 4.
CYP734A50 and GLO2 arose by stepwise duplications. (A) Analysis of linkage between CYP734A51 and GLO1 in a mapping population of P. veris. Colors represent different genotypes as indicated. (B) Relationship of Ks values for comparisons of CYP734A-like sequences and GLO-like sequences for different species pairs (red and orange) and for S-locus to non-S-locus paralogues within the four species P. veris, P. forbesii, P. oreodoxa, and P. maximowiczii. The Ks values on which this plot is based can be found in Table 2. (C) Schematic phylogeny of the species used for the analyses in D and E based on refs. 17, 40, and 67. The species abbreviations used in D and E are given in brackets. (D and E) Phylogenies of CYP734A-like (D) and GLO-like sequences (E). For the Primula species, paralogues located at the S locus are indicated in cyan, while paralogues located outside of the S locus are shown in orange. Numbers on branches indicate bootstrap support from 1,000 runs. Bootstrap values below 50% are not shown.