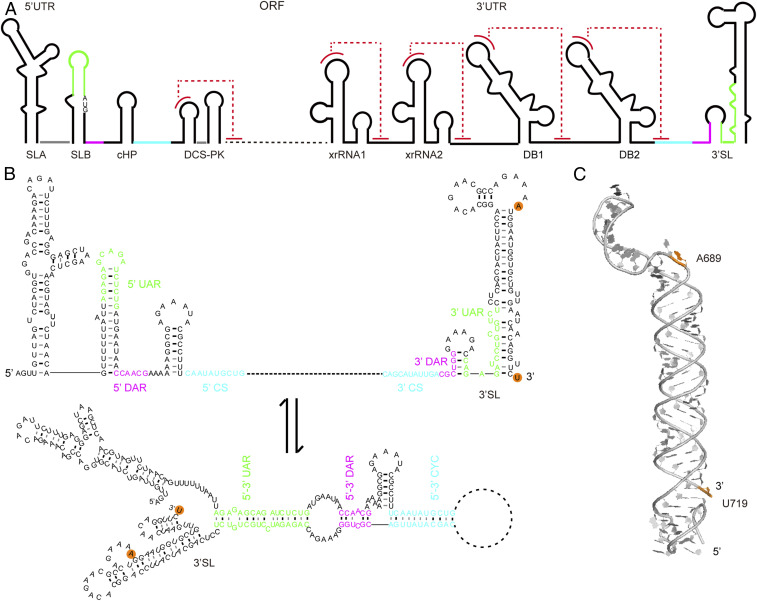
Fig. 2.
Conformational changes in the 3′SL RNA element of DENV2 upon genome cyclization. (A) The single-stranded positive sense genomic RNA of DENV2 consists of an ORF flanked with many highly structured RNA elements, such as the SLA, SLB, cHP, and DCS-PK elements located in the 5′-UTRs and the capsid protein coding regions, the xrRNA1, xrRNA2, DB1, DB2, and 3′SL elements located within the 3′ UTR. Pseudoknot formations in these elements are colored by dashed lines in red. (B) Three sets of long-range RNA–RNA interactions: 5′-3′ upstream of AUG region (UAR) (green), 5′-3′ downstream of AUG region (DAR) (magenta), and 5′-3′ cyclization sequence (CS) (cyan) mediate the linear-to-circular conformational transition of the genomic RNA upon genome cyclization, resulting in significant structural changes in RNA elements, such as the SLB and 3′SL. The modification or labeling sites corresponding to A689 and U719 in DENV-mini are indicated with orange backgrounds. (C) Atomic model of the 97-nt 3′SL element by SAXS and computation, which forms an extended conformation coaxially stacked by the sHP and the long SL (42). The distance between N9 of A689 and N1 of U719 is measured as 83.3 Å in the atomic model.