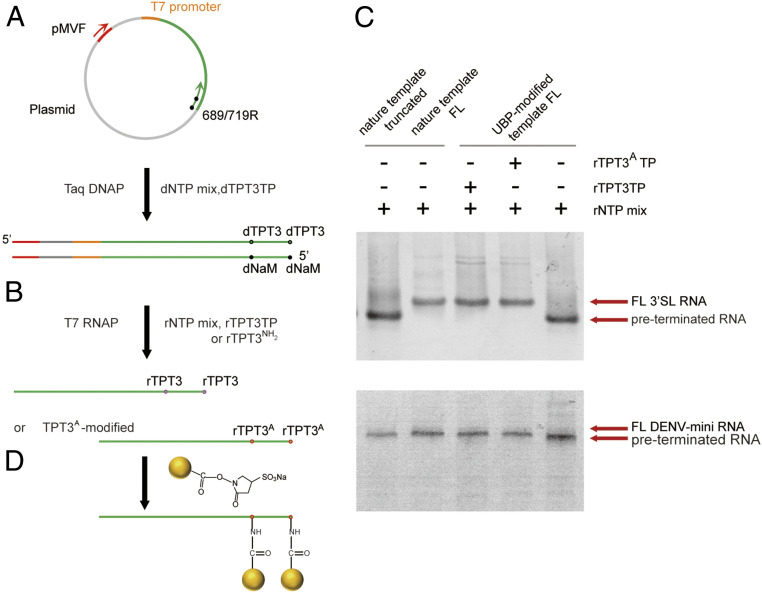
Fig. 3.
A general strategy for site-specific Nanogold labeling of large RNAs using an expanded genetic alphabet containing TPT3-NaM. (A) The UBPs were first incorporated into the reverse ssDNA primers targeting the plasmids by solid-phase chemical synthesis with the phosphoramidites of dNaM or dTPT3. Directed by an upstream forward primer and the respective reverse primers, dsDNA templates containing UBPs at specific sites were amplified by PCR. (B) rTPT3 or rTPT3A can be incorporated into RNAs by in vitro transcription with dsDNA templates containing UBPs (dNaM in the template strand) using the six-letter expanded genetic alphabet. (C) 10% (for 3′SL, top) or 6% (for DENV-mini, down) native PAGE analysis of the in vitro transcripts from natural DNA templates or UBP-modified unnatural DNA templates. The transcription of truncated (up to A688) or full-length (FL) natural DNA template (Left two) using rNTP mix resulted in one single band for 3′SL (top) and DENV-mini (down). The transcription of unnatural DNA template (Right most) in the absence of unnatural nts resulted in preterminated smeared bands. The transcription of the unnatural DNA template (Middle) in the presence of rTPT3 or rTPT3A resulted in a singe dense band with similar migration as FL RNAs. (D) MonoSulfo-NHS-Nanogold labels are coupled to TPT3A-modified RNAs via the amine-NHS ester reaction.