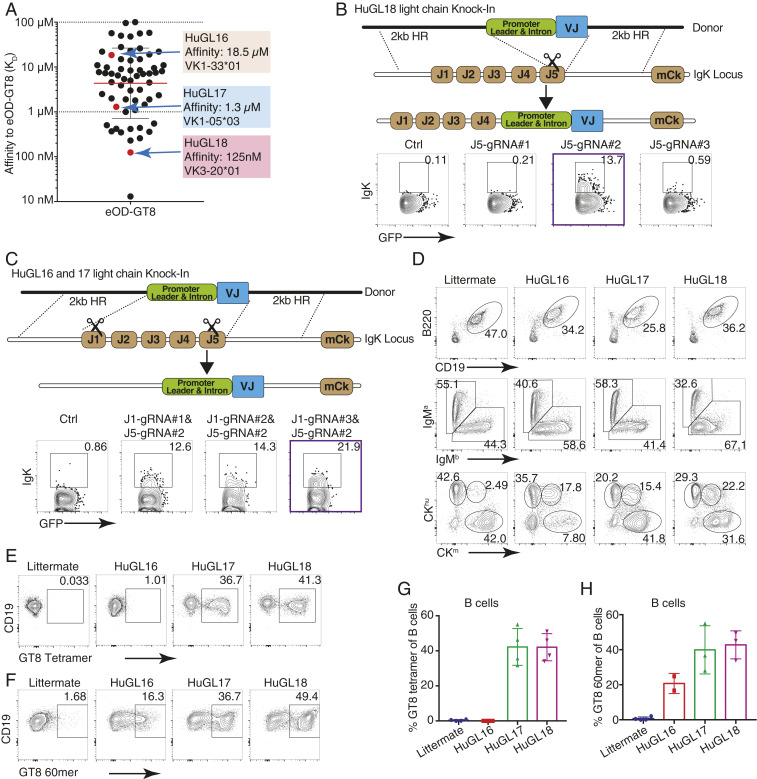
Fig. 1.
Naive human B cell BCR VRC01-class H/L knockin mouse models HuGL16, HuGL17, and HuGL18. Figure shows the design and features of HuGL knockin mice. (A) Affinities of VRC01-class BCRs cloned from HIV– healthy human donors (43) highlighting the KD values and LC usage of HuGL16, HuGL17, and HuGL18. (B and C) HC knockins were generated using a conventional targeting strategy as described (13, 47, 48). LC knockin strategies are shown in the schematics, detailing the CRISPR-facilitated targeting strategy used for HuGL18 (B) or HuGL16 and HuGL17 (C), which involved one or two cuts in the Jκ locus, respectively. HR, arms of homology. Flow plots below each schematic show transient transfection analysis of gene targeting efficiencies in a Rag1−/− pro-B cell line using CRISPR ribonucleoproteins carrying the indicated guide RNAs and LC targeting construct (Materials and Methods). (D–H) Characterization of lymphocytes in HuGL mice. (D) Alleles were marked by breeding BCR knockins of interest (C57BL/6 background: Ighb/bCκm/m) to Igha/aCκhu/hu mice, followed by flow cytometry analysis with antibodies that distinguish C regions. (E–H) Evaluation of antigen-binding by B cells of the indicated strains using eOD-GT8 streptavidin tetramers (E) or eOD-GT8 60mer nanoparticles (F), with quantification shown in G and H. Data are representative of multiple litters per HuGL. (E–H) n = individual experiments, n = mice per group. n = 3. n = 1 to 3 mice per experiment. See also SI Appendix, Fig. S1.