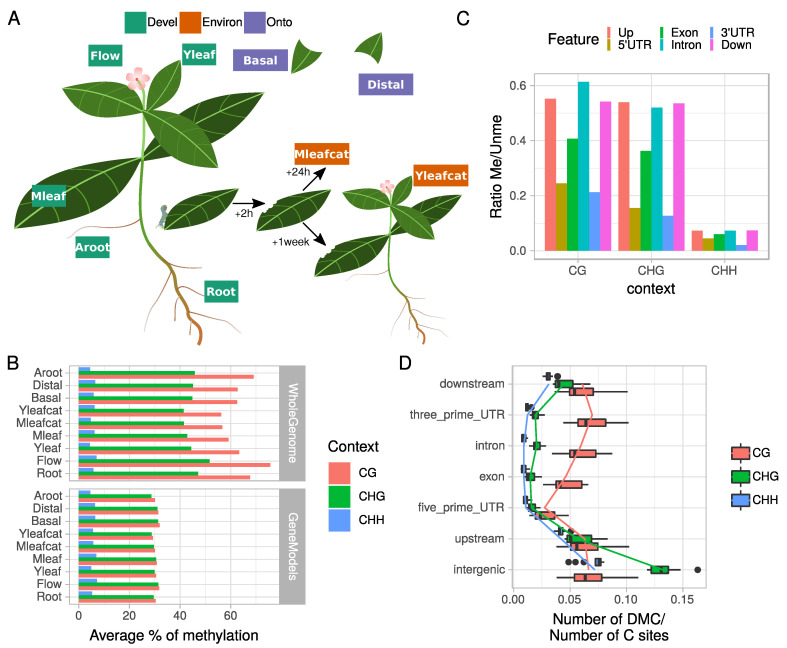
Figure 1.
Overview of theC. roseusexperimental design for the WGBS and methylome data. (A) Nine samples reflecting developmental variation within distinct organs (Leaf, Flower, Root or Adventitious Root) or during leaf ontogenesis (young versus mature; or basal versus distal) and environmental effects on leaf after direct (mature leaf 24 h post folivory) or indirect (distance young leaf 1 week post folivory) biotic attacks were chosen to characterize whole genome cytosine methylation profiles. Flow = Flower; Aroot = Adventitious root; Mleaf = Mature leaf; Yleaf = Young leaf; Basal = Basal part of leaf; Distal = Distal part of leaf; Mleafcat = Mature leaf post folivory by caterpillars; Yleafcat = Young leaf from subjected plants 1 week after folivory on mature leaves. (B) Average % of cytosine methylation in the whole genome or gene model targeted approaches. (C) Average cytosine methylation levels of different gene features in each cytosine context for the whole genome approach (D) ratio between the number of differentially methylated cytosines (DMC) and number of cytosine sites per feature and cytosine contexts (CG, CHG, or CHH). In the whole genome approach, DMCs were retained if the difference in methylation between the two samples considered was >25%.