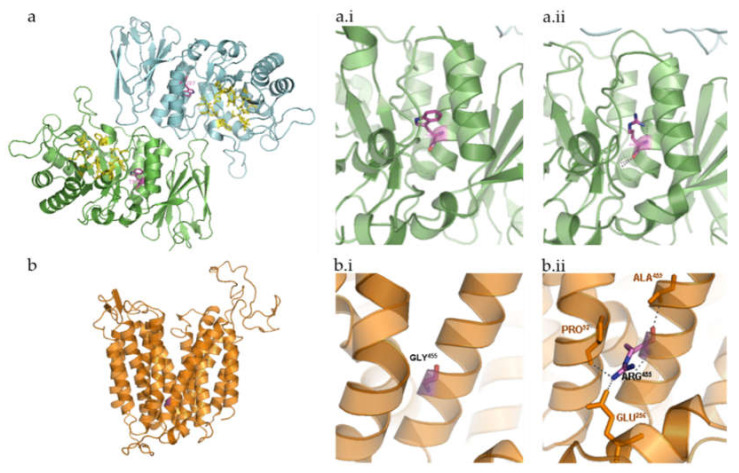
Figure 3.
Three-dimensional structure representation of α-galactosidase (GLA) and major facilitator superfamily domain containing 8 (MFSD8) using PyMol. β-strands, helices, and coils indicate the secondary structure elements that form the scaffold for the interacting residues. Relevant side chains that interact with the residues of interest are depicted and labelled. (a) Human GLA dimer, shown in ribbon representation (Protein Data Bank accession code = 1R46). Each monomer of the homodimer contains two domains, α(β/α)8 barrel containing the active site plus a C-terminal antiparallel β domain. Active site residues are shown in yellow (Trp47, Asp92, Asp93, Tyr134, Cys142, Lys168, Asp170, Glu203, Leu206, Tyr207, Arg227, Asp231, Asp266, Met267). Relevant side chains that interact with the residue of interest are depicted and labelled. α-galactosidase A dimer, with the active site residues and the affected amino acid highlighted; (a.i) wild-type Trp287; (a.ii)—mutated Arg287; (b) in silico design of MFSD8 three-dimensional structure based on sequence analysis and subsequent prediction of the secondary structure elements it encodes. The three-dimensional structure depicted does not take into consideration the fact that this is a multi-pass membrane protein. Instead, it refers to the mature protein MFSD8 as it would coil based solely on the intra-molecular interactions. Still, a comparison between this prediction and that of the transmembrane domains further validates the theoretical assumption that MFSD8 contains 12 transmembrane domains, which are formed by α-helices. (b.i) Wild-type Gly455. (b.ii) Mutated Arg455.