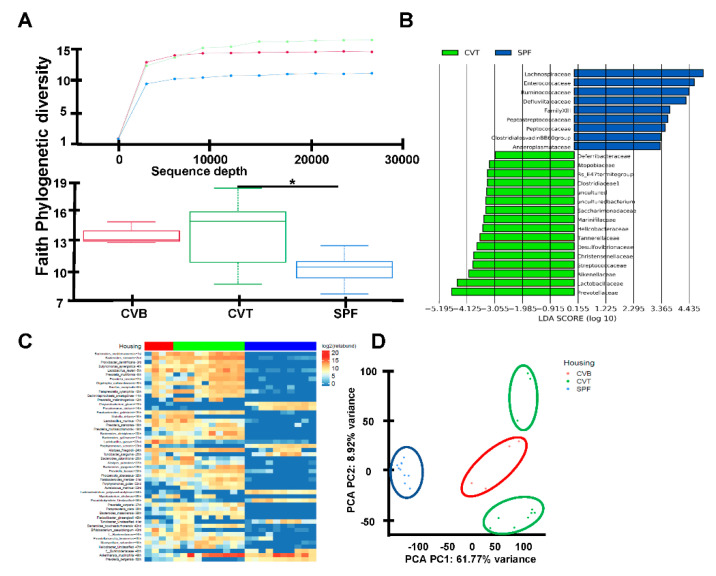
Figure 2.
(A) C57BL/6J (B6) mice born and raised in specific-pathogen-free (SPF) facilities were either maintained in SPF or were transferred to a conventional facility and co-housed (CVT) with mice born in that facility (CVB) for one month. Fecal samples were collected from SPF (n = 18), CVB (n = 3), and CVT (n = 15) mice at one (n = 12) or six to twelve (n = 3) months of co-housing, and were then processed for DNA extraction and 16S rRNA gene amplicon sequencing to assess microbiota phylogenetic diversity, shown as rarefaction plot using the Faith phylogenetic diversity metric for alpha-diversity and box plots showing significant difference (p value = 0.01) in Faith Phylogenetic diversity between CVT and SPF mice. (B) Differentially abundant taxa across CVT and SPF mice are shown as LEFse plot. (C) Fecal DNA samples from CVT (n = 8), SPF (n = 9) and CVB (n = 4) mice were also proceeded for shotgun metagenomics analyses shown as ranked 50 most variant last known taxa differentially represented in CVT, SPF and CVB mice. (D) Display of taxa data based principal components 1 and 2 distribution resulted in specific clusters for CVT, SPF and CVB fecal samples.