Abstract
Understanding how cell organelles and compartments communicate with each other has always been an important field of knowledge widely explored by many researchers. However, despite years of investigations, one point—and perhaps the only point that many agree on—is that our knowledge about cellular-signaling pathways still requires expanding. Chloroplasts and mitochondria (because of their primary functions in energy conversion) are important cellular sensors of environmental fluctuations and feedback they provide back to the nucleus is important for acclimatory responses. Under stressful conditions, it is important to manage cellular resources more efficiently in order to maintain a proper balance between development, growth and stress responses. For example, it can be achieved through regulation of nuclear and organellar gene expression. If plants are unable to adapt to stressful conditions, they will be unable to efficiently produce energy for growth and development—and ultimately die. In this review, we show the importance of retrograde signaling in stress responses, including the induction of cell death and in organelle biogenesis. The complexity of these pathways demonstrates how challenging it is to expand the existing knowledge. However, understanding this sophisticated communication may be important to develop new strategies of how to improve adaptability of plants in rapidly changing environments.
Keywords: retrograde signaling, biogenic control, operational control, stress response, cell death
1. Introduction
Oxygenic photosynthesis is an ancient process that likely evolved over 3.7 billion years ago in free-living bacteria. According to the endosymbiosis theory, the ancestor of the current alphaproteobacteria from which mitochondria are derived were incorporated into prokaryotic Archaean cells. Some of these newly formed eukaryotic cells underwent another endosymbiosis event, incorporating a photosynthetically active ancestor of cyanobacteria into their cells. These two events—followed by a long period of evolution—resulted in the emergence of modern eukaryotic plant cells [1]. During the evolution of eukaryotic plant cells, the genomes of the original endosymbionts evolved and rearranged in such way that many genes were transferred from organellar genomes to nucleus. This process was aimed at securing endosymbionts in eukaryotic cell and simplifying metabolic pathways to allow eukaryotic cells to manage their resources in a more efficient manner. However, these genetic rearmaments during the evolution of the modern plant cell also required the evolution of a communication between the prokaryotic precursors of organelles and the eukaryotic nucleus.
During evolution, the majority of genes encoded by the genome of free-living cyanobacteria were transferred to the nuclear genome of the newly formed symbiotic cell [2,3]. Around 18% of the Arabidopsis thaliana nuclear genes are derived from a plastid ancestor. Surprisingly, the majority of proteins encoded by those genes are not transferred back to the chloroplasts. Even in nonphotosynthetic organisms like Plasmodium or Trypanosoma—which evolved from red algae—we still find some functional genes of cyanobacteria origin [4]. This demonstrate that the benefits from endosymbiosis can be various and the evolution of eukaryotes took many different paths in order to gain advantage over prokaryotic cells. We cannot clearly distinguish which pathway was more beneficial, but it is obvious that the emergence of the eukaryotic cell led to the possibility for the evolution of plant, fungi and animal modern cells, which was a major breakthrough in evolution of oxygenic life on earth. To make this happen, the original symbionts had to develop a highly coordinated two—or three-way communication system. It is obvious that the expression of many genes encoded by the nuclear genome depends on organellar signals, in a mechanism called retrograde signaling. In this review, we present role of the signals derived from dysfunctional organelles such as chloroplasts, mitochondria and peroxisomes in regulation of nuclear gene expression (NGE). The proper folding and assembly of many plastid protein complexes (e.g., photosystems) requires the highly coordinated coupled expression of photosynthesis-associated nuclear genes (PhANGs) and photosynthesis-associated plastid genes (PhAPGs). On the other hand, organellar gene expression (OGE) is controlled by nuclear-encoded factors; this kind of regulation is called anterograde signaling. Disturbances in retrograde—as well as anterograde—signaling may lead to their uncoupled expression, which has harmful effects on plant cells by impairing the proper functioning of chloroplasts [5]. Under photooxidative stress, chloroplasts are not able to carry out the biosynthesis of carbohydrates efficiently, which limits the energy supply of cells. Instead, excess excitation energy (EEE) results in a rapid foliar temperature increase due to nonphotochemical quenching (NPQ) and formation of reactive oxygen species (ROS) which—apart from their destructive capabilities—are also able to act as signaling molecules, allowing plants to adapt to suboptimal environmental conditions. The inhibition of mitochondrial electron transport and the tricarboxylic acid cycle requires retrograde induction of the alternative oxidase pathways in order to reduce oxygen and maintain energy production. Alternative respiratory pathways have a lower ATP yield than the main pathway. However, in stressful conditions, it may be the only way to produce energy. Severe oxidative stress can surpass the abilities of the antioxidative system, and ultimately results in the induction of programmed cell death (PCD). Plants induce PCD in order to survive in extreme environmental conditions, or to prevent spread of pathogens which eventually could lead to death of the whole plant organism. Retrograde signals originated from peroxisomes are connected with the inhibition of catalases. Recently, the role of peroxisome-derived H2O2 in the induction of PCD was established; we will discuss this later in this review. On the other hand—apart from discussing the role of each mentioned organelle in retrograde signaling—we also present several putative signal integrators, indicating that under some conditions, changes in the NGE may require a coordinated signal from more than one type of organelle.
Crosstalk between the nucleus and other organelles is crucial, not only for their development, but also to trigger stress responses or to acclimate to a constantly changing environment. Understanding of those complex signaling pathways may allow us to modify them in the future in order to improve crop productivity or to respond to several combined abiotic and biotic stresses [6,7,8,9].
Along with climate change and the overheating of the Earth which we are experiencing nowadays, extreme conditions such as water deficit, extreme temperatures, storms, deforestation, desert expansion and high soil salinity occurs more often [10,11]. The Pace of those changes raises at least a few questions: Will plants be able to cope with such rapid climate change? What about diseases and pests that will migrate to more favorable habitats? This is currently happening, for example in the pandemic wheat disease—stem rust (TTTTF)—which is migrating from Africa to Europe [12]. This is why we should think about these challenges more carefully in coming years. If not, we will be not prepared for what may come [13]. One strategy to resolve this problem is to enhance natural plant defense and acclimation mechanisms in crops with genes from their wild ancestors. Especially helpful in this case may be some knowledge about retrograde-signaling pathways. However, acclimation to stressful conditions requires energetic investment which could be utilized for growth and development. Finding the proper balance between those two processes may be crucial [14].
Even widely described, retrograde-signaling pathways still has some unknowns (mostly how signals are transduced from organelles to nucleus). Recently, some scientists working in this field question that even genes that has been considered to be key parts of some retrograde-signaling pathways for last few years. In this review, we try to describe different classes of retrograde signals, already known pathways related to organelles biogenesis or stress response and also discuss all the controversy that have recently arisen around some of the putative retrograde candidates.
2. Plurality of Putative Retrograde Signals
Retrograde signals can be classified according to the nature of the signal. We can distinguish few major types of the signals which are transduced by different biomolecules such as: RNA, ROS, proteins and other metabolites. In pioneering work of chloroplast-to-nucleus signaling, it was proposed that RNA from plastids can regulate protein synthesis in cytoplasm [15]. Indeed, recently was identified link between retrograde signaling and RNA editing in chloroplasts [16]. Noteworthy, recent studies show that RNA metabolism processes such as alternative splicing and micro RNA synthesis are not only affected by retrograde signaling but can also trigger it as well. One hypothesis indicates that it could be achieved through alterations in RNA editing levels for transcripts encoding subunits of RNA polymerase such as rpoC1 and rpoB [17,18,19,20]. Even without evidence of RNA being signaling molecule for retrograde signaling itself, it is clear that RNA metabolism (both nuclear and plastid) plays major role in transduction of retrograde signals.
ROS are products of several metabolic pathways; it was described that their accumulation can affect NGE [21]. ROS can be produced in apoplast, chloroplasts, mitochondria, glyoxysomes, peroxisomes, endoplasmic reticulum (ER) and cytosol [22]. Photosynthesis and photorespiration are well known metabolic processes that generates ROS. NADPH oxidases (NOXs) activity results in ROS formation in apoplast. Respiratory burst oxidase homologs (RBOHs) are plant NADPH oxidases located in the plasma membrane. Apoplastic ROS are scavenged in the same way as it happens intracellular, but by a specific extracellular isoforms of antioxidative enzymes [23,24]. H2O2 formed in apoplast can be transported into the cells via aquaporins (AQPs) (Figure 1). Plasma membrane intrinsic protein 1.4 (AtPIP1.4) was described as the AQP that can facility intracellular H2O2 transport through plasma membrane from apoplast [25]. This mechanism may allow apoplastic communication between plant cells and allow induction of systemic responses. Redox state of the apoplast is important in acclimation of photosynthesis to variable light intensity [26]. To further support this hypothesis recently apoplastic H2O2 sensor—HPCA1—was identified [27]. Cells developed ROS scavenging mechanisms to maintain homeostasis, it is achieved mainly by enzymatic and nonenzymatic compounds [28]. Glutathione (GSH) and especially ascorbate (AsA) are main nonenzymatic antioxidants in plants [29]. However, those nonenzymatic compounds depend on their recycling by specific set of enzymes. For example, DEHYDROASCORBATE REDUCTASE2 (DHER2) is an important enzyme taking part in ascorbate recycling [30]. There are also other nonenzymatic antioxidative compounds such as phenolic compounds, carotenoids, flavonoids, tocopherols and alkaloids [31]. Mentioned compounds play role in regulating redox state of plant cells. Under abiotic or biotic stress ROS production surpass abilities of antioxidative systems eventually changing redox state of chloroplasts which is considered to be important in triggering retrograde signaling. Because some of ROS like singlet oxygen (1O2) have short lifespan it is clear that it rather takes part as one of the signaling molecules than migrate from plastids to nucleus itself to modulate NGE. It is still unclear how ROS escape chloroplasts. However, in cytoplasm there are several redox sensitive proteins such as Mitogen-activated protein kinases (MAPKs) which can trigger signal cascades that eventually lead to changes in NGE [32]. ROS stability, apart from their short half-time is also linked with their reactivity. More reactive species like hydroxyl radical (OH●) are chemically unstable and they can only oxidize compounds in their close vicinity. In contrast to hydroxyl radical, H2O2 is the most stable ROS. However, short-lived ROS like superoxide anion radical (O2•−) generated by photosystem I (PSI) can be dismutated to H2O2 enzymatically by superoxide dismutase (SOD) or spontaneously [33]. It was reported that hydrogen peroxide can oxidase Calvin cycle enzymes containing thiol group [34]. Oxidation of cysteine thiol group to sulfenic acid (Cys–SOH) is well established reversible post translational modification that regulates protein activity. It depends on the redox state of cell. Sulfenylation of important antioxidant enzyme DHER2 prevents it from irreversible overoxidation [30]. Endosymbiosis events also contributed to evolution of redox sensitive proteins containing cysteine thiols in order to sense and regulate redox state of cell [35]. Many hypotheses about role of H2O2 in plant signaling under light illumination were thoughtfully discussed in literature [36,37]. H2O2 derived from photosynthesis is also able to diffuse out of the chloroplasts during illumination [32,38,39]. However, authors suggested that it cannot pass chloroplast membrane by simple diffusion. It is more likely that it is transported out of chloroplasts through AQPs [40]. Some researchers hypothesize that it may be delivered from chloroplast to nucleus via stromules [41]. They are tubules filled with stroma formed by all plastid types discovered in vascular plants and it was described that they can emanate after exposure to ROS [42]. Recently, research group of Phillip Mullineaux suggested that photosynthesis derived H2O2 from chloroplasts associated nearby nuclei may affect NGE omitting transport by cytosol [43].
Figure 1.
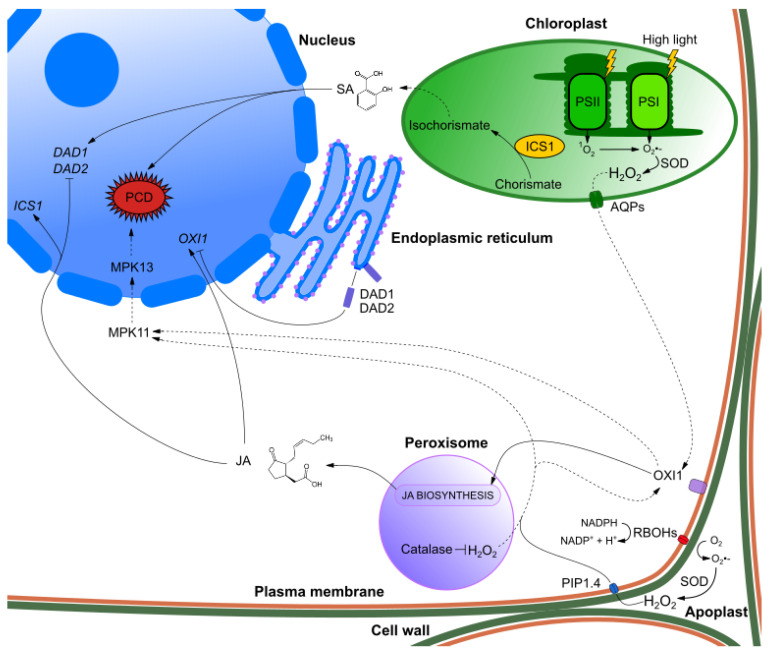
Role of oxidative signal inducible 1 (OXI1) in retrograde signaling in mature mesophyll cells. OXI1 in response to reactive oxygen species (ROS) induces biosynthesis of jasmonic acid (JA) and induces programmed cell death (PCD) through MPK11/MPK13-mediated pathway. JA induces expression of ICS1. Crosstalk between SA and JA along with endoplasmic reticulum (ER)-associated proteins DEFENDER AGAINST CELL DEATH (DAD1)/DAD2 regulates expression of OXI1. Dotted lines represent pathways which lack experimental evidence or exact nature of indicated regulation is still unknown.
Hydrogen peroxide in chloroplasts is detoxified by peroxiredoxin (PrxR), but mostly by ascorbate–glutathione cycle in which ascorbate peroxidase (APX) is key enzyme [44,45]. There are different enzymes that can also scavenge ROS, including catalase (CAT), superoxide dismutase (SOD) and glutathione peroxidase (GPX). Because of their broad substrate specificity usually each ROS can be utilized by at least two different enzymes [46,47]. It is also worth mentioning that each cell compartment has specific protein isoforms and contain more than one type of antioxidative enzymes. For example, we can distinguish APX isoenzymes located in stromal (sAPX) and thylakoid (tAPX) membrane of chloroplasts (in mature mesophyll cells), cytosol APX (cAPX) and microbody APX (mAPX) bound to membrane of peroxisome and glyoxysome [48]. APX change their activity under high light illumination [49,50,51]. APXs expression is regulated by redox status of plastoquinone pool [50]. Existence of several APX isoenzymes allow plant cells to regulate ROS content in each compartment independently [52]. Cells may purposely not scavenge all ROS in efficient manner and in every cell compartment so the signal can be transduced and proper response for high light intensity induced. It is still unclear if response to H2O2 depends on in which organelle it was produced or rather signal is integrated regardless of the source [53]. Signaling pathways originating from peroxisomes were often discovered analyzing catalase mutants (cat) which accumulate H2O2 inside peroxisomes in differentiated cells. Its accumulation lead to the induction of pathogenesis related genes and ultimately to the cell death [54,55]. cat mutants differentially express many nuclear encoded genes compared to the wild-type plants under high light intensity stress [56]. Transcriptome analysis of cat2 mutant shows higher expression of many genes related to protein repair [57]. More severe changes are observed in transcriptome of cat1 cat2 cat3 triple mutant exhibiting deregulation in expression pattern of several receptor-like kinases and transcription factors (TFs) [58]. Among those genes oxidative signal inducible 1 (OXI1) encoding serine/threonine kinase, MPK11 and MPK13 were especially interesting considering redox sensitivity of some MAPKs. OXI1 belongs to AGC family of plasma membrane bound kinases [59]. OXI1 was described as activator of MPK3 and MPK6 in response to H2O2 and pathogens [60]. However, no changes in expression pattern of those kinases were observed in triple catalase mutant [58]. This may suggest existence of separate H2O2 related signaling pathways for peroxisomes and chloroplasts. H2O2 accumulation in peroxisomes promote transduction of the signals through OXI1/MPK11/MPK13 (Figure 1). To support this statement, it was observed that MPK3 and MPK6 expression levels are not affected in OXI1 overexpressing lines [61]. Recently, two PCD inhibitors: DEFENDER AGAINST CELL DEATH (DAD1) and its homolog DAD2 were described as regulators of OXI1 induced cell death in response to high light stress [61]. DAD1 and DAD2 overexpression lines exhibit lower expression of OXI1 than wild-type and OXI1 is upregulated in dad1 and dad2 mutants. This suggest antagonistic role of DAD1/DAD2 and OXI1. Induction of OXI1 dependent cell death is coregulated by two plant hormones salicylic acid (SA) and jasmonic acid (JA). SA apart from its role in pathogen defense signaling also regulates redox homeostasis and light acclimation [62]. Wild-type plants treated with JA exhibit increased expression of OXI1 and decreased expression of DAD1 and DAD2. oxi1 mutants have lower content of JA than wild-type plants and high light treatment does not induce JA accumulation like it does in the wild-type. This suggest that OXI1 regulates JA biosynthesis in peroxisomes. In contrast, treatment with SA induces DAD1 and DAD2 expression while OXI1 is expressed at same level. Treatment with JA induce expression of SA biosynthesis genes: ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and ISOCHORISMATE SYNTHASE1 (ICS1) in the wild-type which leads to increased isochorismate biosynthesis in chloroplasts. ICS1 converts chorismate to isochorismate which is a precursor for SA biosynthesis in cytosol. In contrast treatment with SA does not affect expression of JA biosynthesis genes (AOS and OPR3) in the wild-type [61]. Regulation of PCD by OXI1 in response to highlight and H2O2 accumulation in peroxisomes are shown in Figure 1.
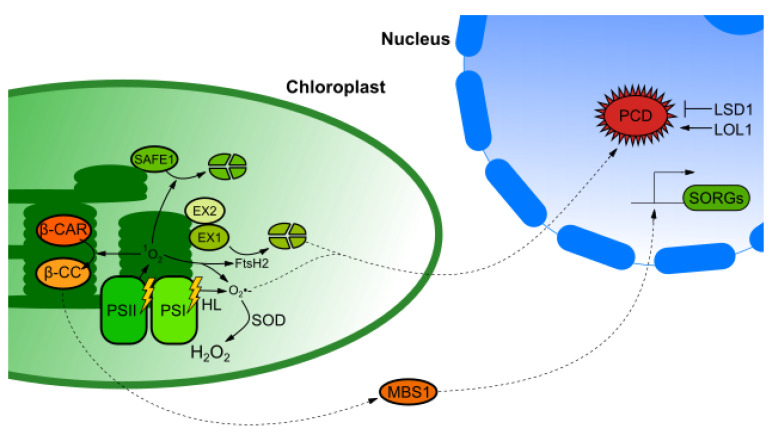
Another interesting aspect in singlet-oxygen–mediated signaling is the executer pathway. EXECUTER1 (EX1) and EXECUTER2 (EX2) are proteins associated with thylakoid membranes and were identified using fluorescent (flu) mutant suppressor screen [63,64]. Seedling of Arabidopsis thaliana flu mutant in the dark accumulate protochlorophyllide which is an intermediate in the biosynthesis of chlorophyll a. Transitioning mutants to light results in burst production of singlet oxygen which affect NGE and eventually leads to the inhibition of growth, chlorosis and ultimately cell death [65]. EX1 and EX2 are part of singlet-oxygen-dependent retrograde signaling under low light conditions [63]. In excess light conditions retrograde signals are transduced independently of EX1 and EX2 through generation of β-cyclocitral [66,67]. EX1 is localized to the grana margins in chloroplasts. Chlorophyll synthesis, disassemble and reassemble of damaged PSII take place in a close vicinity of EX1. Singlet-oxygen–mediated retrograde signaling depends on degradation of EX1 protein by ATP-dependent zinc metalloprotease FtsH. The FtsH also catalyzes cleavage of D1 protein in the reaction center of the damaged PSII. Based on that, it is suggested that EX1 dependent signaling is connected with repair of PSII [68]. EX1 degradation depends on oxidation of tryptophan at position 643 by singlet oxygen. The substitution of this amino acid with leucine or alanine (which also are singlet oxygen sensitive amino acids) inhibits EX1 degradation by FtsH2 [69]. Recently, another novel singlet oxygen induced retrograde singling pathway was discovered by ethyl methanesulfonate (EMS) mutagenization in flu ex1 double mutant. SAFEGUARD1 (SAFE1) is localized in the stroma of chloroplasts and is degraded by the release of singlet oxygen. Plants lacking functional SAFE1 protein were more susceptible to singlet-oxygen-induced damage of thylakoids grana margins [70]. Singlet-oxygen-mediated retrograde-signaling pathways are demonstrated in Figure 2.
Figure 2.
Retrograde-signaling pathways mediated by singlet oxygen in mature mesophyll cell. High light stress induces ROS production in photosystem I (PSI) and PSII. Depending on its concentration singlet oxygen induce different pathways. At low concentrations it promotes degradation of EXECUTER1 (EX1) by FtsH2 protease and induce EX1-mediated PCD. It is negatively regulated by LESION STIMULATING DISEASE1 (LSD1) and positively by its homolog LOL1. SAFEGUARD1 (SAFE1) similarly, to EX1 protects grana margins from oxidative damage by singlet oxygen. At high concentration singlet oxygen oxidizes β-carotene to β-cyclocitral and induces expression of many singlet oxygen related genes (SORGs) through METHYLENE BLUE SENSITIVITY 1 (MBS1). Dotted lines represent pathways which lack experimental evidence or exact nature of indicated regulation is still unknown.
Retrograde signals can also be carried out by different classes of proteins in which transcription factors are worth mentioning. There are few well described proteins such as Whirly1, Plant homeodomain transcription factor with transmembrane domains (PTM) or ABSCISIC ACID-INSENSITIVE4 (ABI4) [71,72,73], but we will describe those widely in other section of this review.
One of most interesting and well known retrograde-signaling pathway is connected with tetrapyrrole biosynthesis. Under norflurazon (NF) treatment—which inhibits biosynthesis of carotenoids—several nuclear genes are downregulated [73,74]. Almost three decades ago research group of Joanne Chory identified several genomes uncoupled (gun) mutants under NF treatment [75]. Arabidopsis thaliana mutants exhibiting gun phenotype are not able to downregulate PhANGs (e.g., LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN1.2 (LHCB1.2)) like wild type plants after chloroplast damage caused by NF treatment [76]. Thus, it was concluded that GUNs are part of retrograde-signaling pathway regulating expression of PhANGs. All GUNs except GUN1 encode proteins involved in tetrapyrrole biosynthesis pathway (TBP) in chloroplasts. GUN1 is pentatricopeptide repeat (PPR) protein localized in chloroplasts containing a small MutS related (SMR) domain. Analysis of gun2-5 mutants initially led to conclusion that Mg–protoporphyrin (MgProto) is a retrograde metabolite able to move between the nucleus and chloroplasts [77]. However, further studies did not support this hypothesis because no correlation was found between level of MgProto and LHCB1.2 expression [78]. In contrary some researchers suggested that this may be due to difficulties with identification of tetrapyrroles by HPLC while their content is low [79]. Recent study confirmed accumulation of MgProto in first two days after newly germinated seedlings were treated with NF. The accumulation of MgProto was also correlated with repression of LHCB1.2 expression suggesting that accumulation of MgProto is a retrograde signal [80].
The identification of gun6 mutant exhibiting higher activity of ferrochelatase 1 (FC1) suggested that heme synthetized by FC1 may be precursor or retrograde signal itself [81]. Overexpression of FC1 targeted to chloroplasts rescued nuclear gene expression after NF treatment and increased expression levels of CA1, LHCB2.1 and GUN4 even without this treatment. However, targeting FC1 to mitochondria did not affect NGE [82]. These results further support role of heme synthetized by FC1 in the chloroplast-to-nucleus retrograde signaling. On the other hand, a link between ROS and tetrapyrroles pathway may be a GUN4 protein. GUN4 and Protoporphyrin IX form 1O2 generating complex which can initiate retrograde signaling [83].
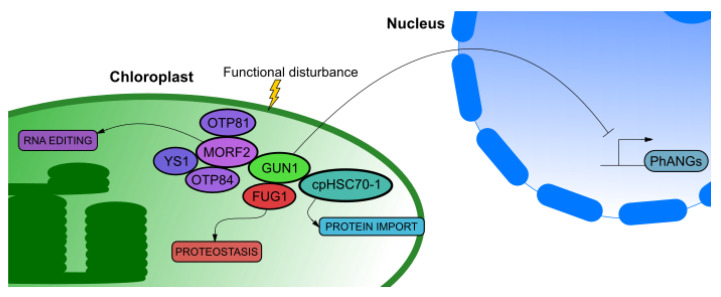
An exception is a GUN1 protein which is not involved in tetrapyrrole synthesis pathway. There are many different hypotheses about exact role of GUN1, but most of them are related to plastid protein homeostasis [84,85,86,87]. When proper functioning of chloroplasts is disturbed GUN1 can also regulate chloroplast RNA editing by physical interaction with MULTIPLE ORGANELLAR RNA EDITING FACTOR 2 (MORF2) to affect maturation of many transcripts among which are subunits of plastid encoded RNA polymerase. MORF2 interacts with ORGANELLE TRANSCRIPT PROCESSING 81 (OTP81), ORGANELLE TRANSCRIPT PROCESSING 84 (OTP84) and YELLOW SEEDLINGS 1 (YS1). otp81, otp84 and ys1 mutants exhibit weak gun phenotype which is enhanced in double and triple mutant in those genes. Overexpression of MORF2 results in strong gun phenotype similar to gun1 mutant suggesting that plastid RNA editing and retrograde signaling are functionally connected [16]. Another GUN1 interacting protein is FUG1 which functions as translation initiation factor in chloroplasts. Both functional proteins are required to maintain plastid protein homeostasis [88]. GUN1 probably does not affect plastid gene expression per se, but it interacts with chaperone cpHSC70-1 in order to regulate nuclear encoded protein import to chloroplast. This allows to maintain protein homeostasis (proteostasis) in the chloroplasts. Mutation in gene encoding cpHSC70-1 leads to a gun phenotype. However, gun1 mutant grown under normal conditions does not have affected protein import capacity [89]. On the other hand, it was recently demonstrated that GUN1 can directly bind to heme as well as other porphyrins, increases FC1 activity and also limits heme and protochlorophyllide synthesis [90]. Based on that, GUN1 is linked to the tetrapyrroles which are considered to be retrograde signaling molecules [76]. Complex role of GUN1 and its interactors were described in Figure 3. Role of transcription factors connected with GUN pathways is described further in this manuscript in a section dedicated to biogenic control.
Figure 3.
Retrograde signals mediated by GENOMES UNCOUPLED 1 (GUN1). In response to functional disturbance of chloroplast GUN1 inhibits expression of photosynthesis-associated nuclear genes (PhANGs), regulates protein import through interaction with cpHSC70-1, maintains proteostasis through interaction with FUG1 and modulates RNA editing through interaction with MULTIPLE ORGANELLAR RNA EDITING FACTOR 2 (MORF2).
An interesting group of metabolites involved in retrograde signaling and mentioned above are carotenoids. They are main scavengers of 1O2 in chloroplasts and products of their oxidation: β-carotene (β-CAR) and its oxidation product β-cyclocitral (β-CC) can induce changes in NGE [91]. However, transduction of the signals through this pathway depends on METHYLENE BLUE SENSITIVITY 1 (MBS1) PROTEIN. MBS1 is a zinc finger protein located in the nucleus and in the cytosol. Lack of functional protein in msb1 mutant caused increased susceptibility to 1O2 generated during high light stress [92]. Increased GFP fluorescence observed in plants expressing MBS1:GFP under native promoter treated with β-CC lead to conclusion that MBS1 is involved in β-CC retrograde-signaling pathway. It is hypothesized that MBS1 can induce expression of singlet oxygen related genes (SORGs) in order to cope with high light intensity stress [93].
Another metabolite linked to retrograde signaling is methylerythritol cyclodiphosphate (MEcPP) [94]. It is a precursor of isoprenoids and its accumulation is corelated with changes in NGE. Increased levels of MEcPP induce unfolded protein response in the ER [95]. Similar to ROS, MEcPP accumulates after abiotic stresses such as wounding or high light [94]. Hydroxymethylbutenyl diphosphate synthase (GcpE) is an enzyme responsible for reducing MEcPP to hydroxymethylbutenyl diphosphate (HMBPP). GcpE is encoded by CEH1 gene. This enzyme is a redox sensitive protein which could explain MEcPP accumulation during photooxidative stress [96]. Recently, two new photoreceptor phytochrome B (phyB) mutant alleles that are able to revert phenotype of constitutively expressing HPL (ceh1) mutant were described. ceh1 mutant was identified in a screen for regulators of stress induced hydroperoxide lyase (HPL) gene [94]. ceh1 mutant exhibits a dwarf phenotype, has high concentration of SA and accumulates MEcPP [97,98].
3′-phosphoadenosine 5′-phosphate (PAP) is another metabolite which can function as a retrograde signaling molecule in response to drought and highlight stress by altering expression of APX2, ELIP2 ZAT10 and DREB2A genes [99]. Abscisic acid (ABA) is one of hormones playing major role in response to those stresses and it is synthetized in chloroplasts. In Arabidopsis thaliana PAP acts as secondary messenger in ABA regulated stomatal closure and germination [100]. PAP is synthetized from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) by sulfotransferases [101]. It was considered to be a byproduct without function in plants however it can alter RNA catabolism in yeast (Saccharomyces cerevisiae) by inhibiting two 5′ → 3′ exoribonucleases (XRNs) [102]. PAP degradation in chloroplasts is mediated by inositol polyphosphate 1-phosphatase (SAL1) which function as nucleotide phosphatase [99]. Point mutation as well as T-DNA insertion in SAL1 gene result in greater drought tolerance which leads to conclusion that SAL1 is a negative regulator of drought tolerance and it is connected with PAP accumulation [103]. Additionally, functional SAL1–PAP pathway is important for biotic stress responses since mutations in SAL1 gene lead to higher susceptibility to Pseudomonas syringae pv. tomato DC3000 and Pectobacterium carotovorum subsp. carotovorum EC1 [104]. SAL1–PAP retrograde-signaling pathway is well conserved in all land plants [105]. It is also interesting that SAL1 is localized in cytosol [106], nucleus [107], chloroplasts [108] and mitochondria [99]. One of most recent reports shows SAL1–PAP retrograde-signaling pathway involvement in iron homeostasis [109].
3. Retrograde Signaling in Regulation of Organelles Biogenesis
Retrograde-pathway-transducing signals from plastids to the nucleus in order to regulate chloroplast biogenesis are often called “biogenic control” [110]. Main purpose of this type of retrograde signaling is to modulate NGE so proteins encoded by several of PhANGs can be produced and transported to chloroplasts during their development from proplastids [111]. The coupled expression of PhANGs and PhAPGs allows proper folding and assembly of photosystem complexes [112]. Disturbances in photosystem stoichiometry leads to photoinhibition because amount of energy absorbed from photons exceeds photochemical efficiency of PSII. Photoinhibition eventually leads to ROS formation which can be lethal for developing seedlings.
Signals conditioning proper plastid biogenesis are connected with tetrapyrrole biosynthesis pathway, changes in plastid gene expression (PGE) and activity of the photosynthetic electron transport (PET). ABI4 was discovered during a screen for ABA-insensitive (abi) mutants which are able to germinate in presence of ABA [113]. Different abi4 alleles were also discovered in independent screens for mutants with altered responses to glucose and other sugars [114,115,116,117]. ABI4 is a TF classified to APETALA2/ethylene-responsive factor (AP2/ERF) family. Genome of Arabidopsis thaliana encodes 147 members of AP2/ERF family and many members of this family are involved in signaling pathways including responses to abiotic and biotic stresses [118,119]. ABI4 takes part in mitochondrial retrograde signaling regulating expression of ALTERNATIVE OXIDASE1a (AOX) and chloroplast retrograde signaling [73,120]. Higher expression of nuclear-encoded RbcS in abi4 mutant compared to the wild-type after NF treatment allowed to conclude that ABI4 is involved in chloroplast retrograde signaling [121]. In addition, it was reported that abi4 mutants were able to rescue expression of LHCB1.2 after lincomycin (Lin) treatment [73]. Lincomycin is plastid translation inhibitor and similar to NF it is often used to screen for gun phonotype. Both of those treatments cause damage to chloroplast resulting in photobleached phenotype and drastically reduced expression of most PhANGs [73,74,122]. Activation of ABI4 depends on phosphorylation by MPK3/MPK6 [123]. Based on that knowledge ABI4 was established as one of key proteins involved in plastid development [71,123,124]. It was considered to be a nuclear target of GUN1-dependent retrograde-signaling pathway [71,73,76]. Although there were also studies in which researchers were not able to observe gun phonotype in abi4 mutant when quantifying expression of CARBONIC ANHYDRASE1 (CA1), GOLDEN2-LIKE1 (GLK2) and LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN1.1 (LHCB1.1) [125,126,127]. In contrast to gun1 mutant, crossing abi4 mutant with plastid protein import2 (ppi2) mutant did not rescue loss of NGE [128]. These results suggest that ABI4 may act independently from GUN1. Recently, an independent study performed on four different alleles of abi4 did not support a role of ABI4 in biogenic retrograde signaling and researchers were unable to obtain strong or consistent gun phonotype in tested abi4 mutant alleles [129].
It is interesting that this is not first gun ‘dismantled’ by this research group. Before, focusing on ABI4 they decided to investigate role of PTM in biogenic retrograde signaling [130]. PTM is a plant homeodomain (PHD) transcription factor bound to chloroplast envelope [71]. It was proposed that PTM can be cleaved off chloroplast membrane after changes in plastid metabolism and its N terminal domain can affect NGE after its accumulation in nucleus [71]. ABI4 is one of genes which expression is induced by PTM [71]. Treatments with Lin and NF lead to conclusion that ptm mutant exhibit gun phenotype because it was able to rescue expression of LHCB after treatment with these chemicals. Since its discovery, PTM has been included in many models describing biogenic control pathways [112,131,132,133]. PTM was also further described in literature by the same research group which provided the first report as regulator of flowering after exposure to high light and it takes part in integration of the signals during de-etiolation [134,135]. However, observation of gun phenotype in ptm mutant was still to be confirmed by another research group. Because of potential important role of PTM in retrograde signaling Matthew Terry group decided to further examine its role under NF and Lin treatment. In conducted experiments they were unable to confirm gun phenotype after both Lin and NF treatment. Expression of selected PhANGs in ptm mutant after treatment were not elevated compared to the wild-type plants. Authors concluded that PTM should be excluded from existing models describing plastid signaling or at least its role in it is not as important as we thought before [130].
There are also other interesting mutants in genes encoding transcription factors that exhibit gun or gun-like phenotype such as: hy5 and glk1glk2 [136,137]. In contrast to the gun phenotype, there are also number of mutants called happy on norflurazon (hon) that can tolerate higher concentration of NF in comparison to the wild-type plants. One of the identified hon mutants had mutation in ClpR4 (HON5) subunit of Clp protease complex which localizes in chloroplasts. Another example is a hon23 mutant, which has mutation in putative chloroplast translation elongation factor, and it clearly shows that these mutations interfere with chloroplast protein homeostasis [138]. It is also worth mentioning that not only mutants in genes encoding transcription factors, but also other proteins such as cry1 (encoding blue light photoreceptor) or coe1 (encoding mitochondrial transcription termination factor 4) can exhibit gun phenotype and eventually take part in biogenic control [137,139].
4. Retrograde Signaling in Stress Response and Acclimation
Retrograde-pathway-transducing signals from plastids to the nucleus in order to cope with environmental stresses and acclimate to them are often called “operational control”. Since plants are in general immobile and unable to avoid many unfavorable environmental conditions, they had to evolve sophisticated mechanisms in order to survive and effectively reproduce.
Chloroplasts are sensors of visible light and crop yield is often corelated with the efficiency of photosynthesis. Second, but equally important metabolic process that provides energy for plant cell is aerobic respiration. This processes however are sensitive to changes in plant growth environment [140]. Abiotic stresses can result in photoinhibition of PSII and inhibition of carbon assimilation enzymes [141]. One of the ultimate responses to severe abiotic and biotic stresses is PCD. Apart from its major contribution during tissue development, PCD is also a mechanism that allows plants to prevent pathogens from reproducing and spreading to uninfected cells. During abiotic stresses PCD promotes dismantling of a limited number of affected cells to prevent severe systemic damage to the whole organism [142]. Cells undergoing PCD exhibit extensive chromatin condensation and developmental PCD can occur only in specific cell types [143]. One of the most interesting regulators of PCD is LESION STIMULATING DISEASE1 (LSD1). LSD1 acts as a transcription regulator and condition dependent scaffold protein [7,144]. LSD1 is negative regulator of cell death and defense responses. Along with its homolog LOL1 (which exhibits an antagonistic function) they cooperate in order to induce adequate response through PCD. Mutation in lsd1 results in a runaway cell death (RCD) in a light dependent manner [145,146]. It was proved that reduction of the PSII antenna size, thus reduction of light absorption and EEE pressure by crossing lsd1 mutant with cao1 mutant caused an increase of NPQ and reversion of the RCD phenotype in lsd1 [147]. The reduction of plastoquinone (PQ) pool induced by EEE results in burst of ROS which lead to induction of SA and ethylene dependent signaling pathway through EDS1 and PHYTOALEXIN DEFICIENT4 (PAD4) which lie on the same pathway as LSD1 because RCD phenotype of lsd1 mutant is abolished by inactivation of ROS, SA and ethylene signaling components [6,147,148]. The overexpression of bacterial salicylate hydroxylase (NahG) fused with chloroplast transit peptide from RbcS in lsd1 mutant background also abolished RCD indicating correlation between lsd1 RCD and SA accumulation. Mutation in lsd1 causes uncoupled expression of PhANGs and PhAPGs before induction of RCD; it was hypothesized that LSD1 can act downstream of GUN-mediated pathway. However, lsd1 treatment with Lin does not rescue expression of LHCBs which indicates that it is not a gun mutant. Based on that knowledge LSD1 is more likely involved in operational than biogenic control. Uncoupled expression of photosynthesis-associated genes in lsd1 cause accumulation of singlet oxygen which result in induction of EX1-mediated PCD. lsd1;ex1 double mutant partially reverts RCD phenotype [5]. Based on that knowledge it is suggested that LSD1 plays a role in regulation of PCD and responses towards biotic and abiotic stresses, it also may be an integrator of SA, ROS (including singlet oxygen), ethylene and other hormones (e.g., IAA) mediated pathways [6]. It is also worth mentioning that LSD1, EDS1 and PAD4 are conditional dependent PCD regulators. For example, in optimal laboratory conditions lsd1 mutant displays deregulation of over 2000 genes while in the suboptimal field conditions it has 62 deregulated genes and only 43 of those genes were commonly deregulated in both of these conditions [9].
In this manuscript, we mainly focused on retrograde pathways related to functioning of chloroplasts. On the other hand, mitochondria also depend on retrograde signaling during their biogenesis and stress responses. However, it needs further investigation whether chloroplasts and mitochondria induce separate signaling pathways or they converge into the same pathways [149]. Knowing that functioning of both is strongly connected through the energy, metabolism and redox status makes second hypothesis a viable one [150,151,152]. However, it is considered that mitochondrial retrograde signaling plays greater role in nonphotosynthetic tissues. To support this hypothesis, it was observed that overexpression of TF ANAC013 in Arabidopsis thaliana resulted in enhanced tolerance of chloroplasts during oxidative stress [153]. It is achieved mostly by dissipating excess of reducing equivalents [154,155,156]. Regulators of mitochondrial retrograde signaling are often identified in genetic screens for TFs that can regulate the expression of nuclear genes encoding mitochondrial proteins related to alternative respiration or stress response. Promoters of AOX1a, UPREGULATED BY OXIDATIVE STRESS (UPOX), NAD(P)H-UBIQUINONE OXIDOREDUCTASE B2 (NDB2) and CYTOCHROME BC1 SYNTHASE1 (AtBCS1) are often used in such screens. After high light and antimycin A (electron transport chain blocker similar to cyanide) treatment, AtWRKY40 downregulated and AtWRKY63 upregulated expression of AOX1a, UPOX, NDB2 and AtBCS1 suggesting their antagonistic function [157]. Interestingly among identified AOX regulators there are many components involved in auxin signaling [158]. Another interesting regulator of mitochondrial retrograde signaling is OM66. OM66 encodes mitochondrial outer membrane protein and its promoter is highly induced by SA in contrary to promoter of AOX1a which is responsive to H2O2 and rotenone [159]. Because PATHOGENESIS-RELATED1 (PR1) is downregulated in OM66 mutant and OM66 overexpressing lines have higher content of SA it was proposed that OM66 is regulated in a SA dependent manner [157]. SA inhibits both cytochrome and alternative respiratory pathways [160]. It also inhibits alpha-ketoglutarate dehydrogenase (α-kGDH) a tricarboxylic acid cycle (TCA) enzyme [161]. This information along with well-established SA interactions with chloroplasts metabolism suggests that SA may be a link between chloroplast and mitochondrial retrograde signaling [162].
Retrograde signals transduced from mitochondria as well as chloroplasts can change NGE in a similar manner. The convergence of those two retrograde-signaling pathways is often linked to CYCLIN DEPENDENT KINASE E1 (CDKE1). It is encoded by REGULATOR OF ALTERNATIVE OXIDASE1 (RAO1) gene. It was first described as an important component of mitochondrial retrograde signaling in response to inhibitors. Functional kinase is needed to regulate AOX1a in response to oxidative (H2O2) and cold stress [163]. It was also demonstrated that CDKE1 regulates expression of AOX1a and Lhcb2.4 in response to photosynthesis inhibitors such as 2,5-dibromo-3-methyl-6-isopropyl-benzoquinone (DBMIB) and 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). cdke1-mutants under high light stress exhibited gun-like phenotype [164]. Based on that it was proposed that CDKE1 can integrate retrograde signals from mitochondria and chloroplasts. KIN10 is a subunit of SnRK1 kinase complex conditioning its catalytic activity and it was proposed as an integrator of stress and energy signaling [165,166]. The interaction between KIN10 and CDKE1 in nucleus was also reported [163]. CDKE1 is a part of mediatory complex regulating RNA polymerase II (RNAP II) dependent transcription. Such complexes are considered to integrate stress signals from organelles and initiate proper transcriptional response [167]. Another similarity can be observed in how mitochondrial retrograde signaling is triggered by stress or dysfunction affecting respiratory electron transport chain or TCA [168]. In order to maintain these processes and their energy production, plants need to change their metabolism. That is why proper communication between organelles and nucleus is required [14].
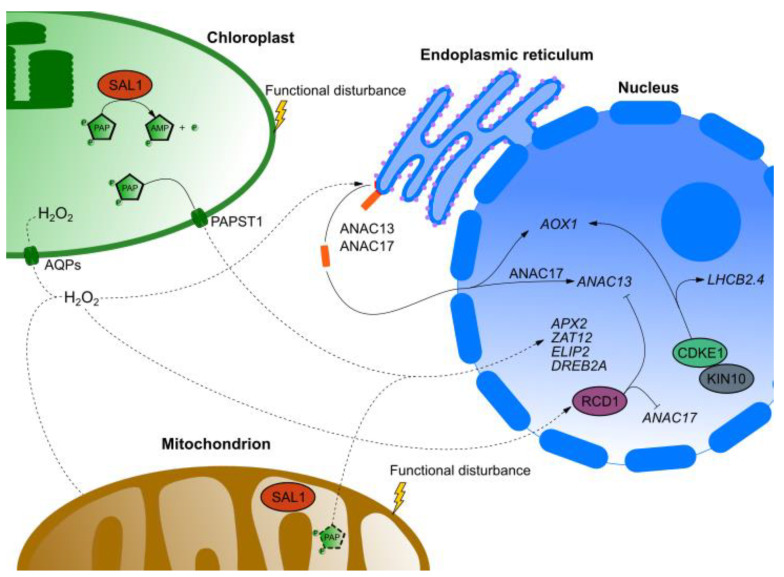
Transcriptome meta-analyses demonstrate that 10% to 20% from differentially regulated genes during abiotic stress responses encode proteins localized to the chloroplasts [169]. We briefly described the role of ROS in retrograde signaling in previous sections of this manuscript. However, it is worth mentioning that ROS signaling is crucial to respond to several abiotic stresses such as drought, variable high light intensity, salinity and heat [170]. Exposure to abiotic and biotic stresses can induce unfolding protein response in ER. As was mentioned before it is connected with MEcPP accumulation [171]. Response to drought stress is usually connected to the SAL1–PAP pathway. Some researchers hypothesize that chloroplasts and mitochondrial retrograde signals converge through TF called ANAC017 (encoded by RAO2) to regulate PCD as response to severe organellar stress [172]. NAC family members are ER bound TFs which upon activation are cleaved and relocated to nucleus where they can affect NGE [173]. RADICAL-INDUCED CELL DEATH1 (RCD1) is another putative integrator of chloroplastic and mitochondrial ROS signaling pathways. It was identified in a screen for sensitivity to ozone [174]. rcd1 mutant is resistant to methyl viologen (MV) and UV-B which suggests that RCD1 may be a ROS sensitive protein [175]. In the rcd1 mutant more than 400 genes are differently expressed under standard growth conditions. Among those genes there are those encoding mitochondrial AOXs as well as chloroplast 2-Cys peroxiredoxin (2CP) [176,177,178,179]. RCD1 interacts with several TFs such as ANAC017 and DREB2A [176,180]. Cleavage of ANAC017 from ER is probably dependent on elevated H2O2 levels [181]. Recently, RCD1 was proposed to act as negative regulator of ANAC013 and ANAC017 and thus integrator of NAC and PAP retrograde-signaling pathways [182]. Putative integrators of retrograde-signaling pathways and their interactors were demonstrated on Figure 4. Recently, β-cyclocitral induced protein SCARECROW LIKE14 (SCL14) was described along with TF ANAC102 and xenobiotic detoxification enzymes in lowering levels of toxic carbonyls and peroxides in order to limit damage to the intracellular components caused by photooxidative stress [183].
Figure 4.
Putative integrators of retrograde-signaling pathways. Inhibition of inositol polyphosphate 1-phosphatase (SAL1) results in accumulation of 3′-phosphoadenosine 5′-phosphate (PAP) which induces expression of many drought and high light related genes such as APX2, ZAT12, ELIP2 and DREB2A. RCD1 after induction with ROS inhibits expression of ANAC13 and ANAC17 which regulates AOX1 expression. CDKE1 induces expression of AOX1 and LHCB2.4 and many other genes as part of mediatory complex regulating RNAPII dependent transcription. Dotted lines represent pathways which lack experimental evidence or exact nature of indicated regulation is still unknown.
5. Conclusions
We briefly demonstrated complexity and different nature of retrograde-signaling pathways. It is commonly considered that in each retrograde pathway there are at least two different types of biomolecules involved. It is worth remembering that the signaling pathways we described are not universal for every cell type and in every epigenetic status. While some of them occur only during development, others such as singlet-oxygen–mediated pathways, can occur only in differentiated, photosynthetically active mesophyll cells. Many existing, even well understood pathways still have unknowns and expanding knowledge about them often brings up more new questions than answers. However, even if we are not yet close to understanding retrograde signaling, benefits from it could be worth the effort. Recently, few interesting strategies to improve crop production under field conditions were demonstrated. First aimed at improvement of photorespiration by implementing synthetic glycolate metabolic pathways into tobacco chloroplasts [184]. Others improved crop production and water-use efficiency by accelerating recovery from photoprotection. It was achieved by combined overexpression of PsbS and genes encoding xanthophyll cycle enzymes [185,186]. In the future studies we shall also consider other physical retrograde signaling pathways, for example, direct heat radiation and vibration of organelles, electrical and calcium wave signaling from chloroplasts and mitochondria.
Acknowledgments
In this section you can acknowledge any support given which is not covered by the author contribution or funding sections. This may include administrative and technical support or donations in kind (e.g., materials used for experiments).
Abbreviations
| 1O2 | Singlet oxygen |
| ABA | Abscisic acid |
| ABI4 | ABSCISIC ACID-INSENSITIVE4 |
| AOX | Alternative oxidase 1a gene |
| APX | Ascorbate peroxidase |
| AQPs | Aquaporins |
| CAT | Catalase |
| CDKE1 | CYCLIN DEPENDENT KINASE E1 |
| DAD | DEFENDER AGAINST CELL DEATH |
| EDS1 | ENHANCED DISEASE SUSCEPTIBILITY1 |
| EEE | Excess excitation energy |
| EMS | Ethyl methanesulfonate |
| ER | Endoplasmic reticulum |
| EX1 | EXECUTER1 |
| EX2 | EXECUTER2 |
| FC1 | Ferrochelatase 1 |
| GcpE | Hydroxymethylbutenyl diphosphate synthase |
| GPX | Glutathione peroxidase |
| GSH | Glutathione |
| GUN | Genomes uncoupled |
| H2O2 | Hydrogen peroxide |
| IAA | Indole-3-acetic acid |
| ICS1 | ISOCHORISMATE SYNTHASE1 |
| JA | Jasmonic acid |
| LHCB | LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN gene |
| Lin | Lincomycin |
| LSD1 | LESION STIMULATING DISEASE1 |
| MAPK | Mitogen-activated protein kinase |
| MBS1 | METHYLENE BLUE SENSITIVITY 1 |
| MEcPP | Methylerythritol cyclodiphosphate |
| MgProto | Mg–protoporphyrin |
| MORF2 | Multiple organellar RNA editing factor 2 |
| NF | Norflurazon |
| NGE | Nuclear gene expression |
| NOXs | NADPH oxidases |
| NPQ | Nonphotochemical quenching |
| O2•- | Superoxide anion radical |
| OGE | Organellar gene expression |
| OH● | Hydroxyl radical |
| OXI1 | Oxidative signal inducible 1 |
| PAD4 | Phytoalexin deficient4 |
| PAP | 3′-phosphoadenosine 5′-phosphate |
| PCD | Programmed cell death |
| PET | Photosynthetic electron transport |
| PGE | Plastid gene expression |
| PhANGs | Photosynthesis-associated nuclear genes |
| PhAPGs | Photosynthesis-associated plastid genes |
| PrxR | Peroxiredoxin |
| PSI | Photosystem I |
| PSII | Photosystem II |
| PTM | Plant homeodomain transcription factor with transmembrane domains |
| RbcS | RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE SMALL SUBUNIT gene |
| RBOH | Respiratory burst oxidase homologs |
| RCD1 | Radical-induced cell death1 |
| RNAP II | RNA polymerase II |
| ROS | Reactive oxygen species |
| SA | Salicylic acid |
| SAFE1 | Safeguard1 |
| SAL1 | Inositol polyphosphate 1-phosphatase |
| SOD | Superoxide dismutase |
| SORGs | Singlet oxygen related genes |
| TBP | Tetrapyrrole biosynthesis pathway |
| TCA | Tricarboxylic acid cycle |
| TF | Transcription factor |
| β-CAR | β-carotene |
| β-CC | β-cyclocitral |
Author Contributions
Conceptualization, J.M., P.G. and S.K.; writing—original draft preparation, J.M.; writing—review and editing, J.M., P.G. and S.K.; visualization, J.M.; supervision, P.G. and S.K.; project administration, S.K.; funding acquisition, S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC was funded by National Science Center in Poland, grant number UMO-2018/29/B/NZ3/01198.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gray M.W. Lynn Margulis and the endosymbiont hypothesis: 50 years later. Mol. Biol. Cell. 2017;28:1285–1287. doi: 10.1091/mbc.e16-07-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stiller J.W. Plastid endosymbiosis, genome evolution and the origin of green plants. Trends Plant Sci. 2007;12:391–396. doi: 10.1016/j.tplants.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Archibald J.M. Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 2015;25:R911–R921. doi: 10.1016/j.cub.2015.07.055. [DOI] [PubMed] [Google Scholar]
- 4.Raven J.A., Allen J.F. Genomics and chloroplast evolution: What did cyanobacteria do for plants? Genome Biol. 2003;4:209. doi: 10.1186/gb-2003-4-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lv R., Li Z., Li M., Dogra V., Lv S., Liu R., Lee K.P., Kim C. Uncoupled expression of nuclear and plastid photosynthesis-associated genes contributes to cell death in a lesion mimic mutant. Plant Cell. 2019;31:210–230. doi: 10.1105/tpc.18.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mühlenbock P., Szechyńska-Hebda M., Płaszczyca M., Baudo M., Mullineaux P.M., Parker J.E., Karpińska B., Karpińskie S. Chloroplast signaling and lesion simulating disease1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell. 2008;20:2339–2356. doi: 10.1105/tpc.108.059618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wituszyńska W., Szechyńska-Hebda M., Sobczak M., Rusaczonek A., Kozlowska-Makulska A., Witoń D., Karpiński S. LESION SIMULATING DISEASE 1 and enhanced disease susceptibility 1 differentially regulate UV-C-induced photooxidative stress signalling and programmed cell death in Arabidopsis thaliana. Plant Cell Environ. 2015;38:315–330. doi: 10.1111/pce.12288. [DOI] [PubMed] [Google Scholar]
- 8.Szechyńska-Hebda M., Kruk J., Górecka M., Karpińska B., Karpiński S. Evidence for light wavelength-specific photoelectrophysiological signaling and memory of excess light episodes in Arabidopsis. Plant Cell. 2010;22:2201–2218. doi: 10.1105/tpc.109.069302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wituszyńska W., Ślesak I., Vanderauwera S., Szechyńska-Hebda M., Kornaś A., Van Der Kelen K., Mühlenbock P., Karpińska B., Maćkowski S., Van Breusegem F., et al. Lesion simulating disease1, enhanced disease susceptibility1, and phytoalexin deficient4 conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiol. 2013;161:1795–1805. doi: 10.1104/pp.112.208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sterling S.M., Ducharne A., Polcher J. The impact of global land-cover change on the terrestrial water cycle. Nat. Clim. Chang. 2013;3:385–390. doi: 10.1038/nclimate1690. [DOI] [Google Scholar]
- 11.Fedoroff N.V., Battisti D.S., Beachy R.N., Cooper P.J.M., Fischhoff D.A., Hodges C.N., Knauf V.C., Lobell D., Mazur B.J., Molden D., et al. Radically rethinking agriculture for the 21st century. Science. 2010;327:833–834. doi: 10.1126/science.1186834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharya S. Deadly new wheat disease threatens Europe’s crops. Nature. 2017;542:145–146. doi: 10.1038/nature.2017.21424. [DOI] [PubMed] [Google Scholar]
- 13.Franks S.J., Weber J.J., Aitken S.N. Evolutionary and plastic responses to climate change in terrestrial plant populations. Evol. Appl. 2014;7:123–139. doi: 10.1111/eva.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baena-González E. Energy signaling in the regulation of gene expression during stress. Mol. Plant. 2010;3:300–313. doi: 10.1093/mp/ssp113. [DOI] [PubMed] [Google Scholar]
- 15.Bradbeer J.W., Atkinson Y.E., Börner T., Hagemann R. Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesised RNA. Nature. 1979;279:816–817. doi: 10.1038/279816a0. [DOI] [Google Scholar]
- 16.Zhao X., Huang J., Chory J. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl. Acad. Sci. USA. 2019;116:10162–10167. doi: 10.1073/pnas.1820426116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godoy Herz M.A., Kubaczka M.G., Brzyżek G., Servi L., Krzyszton M., Simpson C., Brown J., Swiezewski S., Petrillo E., Kornblihtt A.R. Light Regulates Plant Alternative Splicing through the Control of Transcriptional Elongation. Mol. Cell. 2019;73 doi: 10.1016/j.molcel.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Fang X., Zhao G., Zhang S., Li Y., Gu H., Li Y., Zhao Q., Qi Y. Chloroplast-to-Nucleus Signaling Regulates MicroRNA Biogenesis in Arabidopsis. Dev. Cell. 2019;48 doi: 10.1016/j.devcel.2018.11.046. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X., Huang J., Chory J. Unraveling the Linkage between Retrograde Signaling and RNA Metabolism in Plants. Trends Plant Sci. 2020;25:141–147. doi: 10.1016/j.tplants.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Petrillo E., Godoy Herz M.A., Fuchs A., Reifer D., Fuller J., Yanovsky M.J., Simpson C., Brown J.W.S., Barta A., Kalyna M., et al. A chloroplast retrograde signal regulates nuclear alternative splicing. Science. 2014;344:427–430. doi: 10.1126/science.1250322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apel K., Hirt H. Reactive oxygen species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 22.Mignolet-Spruyt L., Xu E., Idänheimo N., Hoeberichts F.A., Mühlenbock P., Brosche M., Van Breusegem F., Kangasjärvi J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016;67:3831–3844. doi: 10.1093/jxb/erw080. [DOI] [PubMed] [Google Scholar]
- 23.Podgórska A., Burian M., Szal B. Extra-cellular but extra-ordinarily important for cells: Apoplastic reactive oxygen species metabolism. Front. Plant Sci. 2017;8:1353. doi: 10.3389/fpls.2017.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sagi M., Fluhr R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 2006;141:336–340. doi: 10.1104/pp.106.078089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian S., Wang X., Li P., Wang H., Ji H., Xie J., Qiu Q., Shen D., Dong H. Plant aquaporin AtPIP1; 4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 2016;171:1635–1650. doi: 10.1104/pp.15.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karpinska B., Zhang K., Rasool B., Pastok D., Morris J., Verrall S.R., Hedley P.E., Hancock R.D., Foyer C.H. The redox state of the apoplast influences the acclimation of photosynthesis and leaf metabolism to changing irradiance. Plant Cell Environ. 2018;41:1083–1097. doi: 10.1111/pce.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu F., Chi Y., Jiang Z., Xu Y., Xie L., Huang F., Wan D., Ni J., Yuan F., Wu X., et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature. 2020;578:577–581. doi: 10.1038/s41586-020-2032-3. [DOI] [PubMed] [Google Scholar]
- 28.Scandalios J.G. Oxidative stress: Molecular perception and transduction of signals triggering antioxidant gene defenses. Braz. J. Med. Biol. Res. 2005;38:995–1014. doi: 10.1590/S0100-879X2005000700003. [DOI] [PubMed] [Google Scholar]
- 29.Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waszczak C., Akter S., Eeckhout D., Persiau G., Wahni K., Bodra N., Van Molle I., De Smet B., Vertommen D., Gevaert K., et al. Sulfenome mining in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2014;111:11545–11550. doi: 10.1073/pnas.1411607111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gratão P.L., Polle A., Lea P.J., Azevedo R.A. Making the life of heavy metal-stressed plants a little easier. Funct. Plant Biol. 2005;32:481. doi: 10.1071/FP05016. [DOI] [PubMed] [Google Scholar]
- 32.Dietz K.J., Turkan I., Krieger-Liszkay A. Redox-and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 2016;171:1541–1550. doi: 10.1104/pp.16.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laloi C., Stachowiak M., Pers-Kamczyc E., Warzych E., Murgia I., Apel K. Cross-talk between singlet oxygen-and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2007;104:672–677. doi: 10.1073/pnas.0609063103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaiser W.M. Reversible inhibition of the calvin cycle and activation of oxidative pentose phosphate cycle in isolated intact chloroplasts by hydrogen peroxide. Planta. 1979;145:377–382. doi: 10.1007/BF00388364. [DOI] [PubMed] [Google Scholar]
- 35.Woehle C., Dagan T., Landan G., Vardi A., Rosenwasser S. Expansion of the redox-sensitive proteome coincides with the plastid endosymbiosis. Nat. Plants. 2017;3:17066. doi: 10.1038/nplants.2017.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullineaux P., Karpinski S. Signal transduction in response to excess light: Getting out of the chloroplast. Curr. Opin. Plant Biol. 2002;5:43–48. doi: 10.1016/S1369-5266(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 37.Mullineaux P.M., Karpinski S., Baker N.R. Spatial dependence for hydrogen peroxide-directed signaling in light-stressed plants. Plant Physiol. 2006;141:346–350. doi: 10.1104/pp.106.078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mubarakshina M.M., Ivanov B.N., Naydov I.A., Hillier W., Badger M.R., Krieger-Liszkay A. Production and diffusion of chloroplastic H2O2 and its implication to signalling. J. Exp. Bot. 2010;61:3577–3587. doi: 10.1093/jxb/erq171. [DOI] [PubMed] [Google Scholar]
- 39.Dietz K.J., Mittler R., Noctor G. Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol. 2016;171:1535–1539. doi: 10.1104/pp.16.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bienert G.P., Møller A.L.B., Kristiansen K.A., Schulz A., Møller I.M., Schjoerring J.K., Jahn T.P. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 2007;282:1183–1192. doi: 10.1074/jbc.M603761200. [DOI] [PubMed] [Google Scholar]
- 41.Caplan J.L., Kumar A.S., Park E., Padmanabhan M.S., Hoban K., Modla S., Czymmek K., Dinesh-Kumar S.P. Chloroplast Stromules Function during Innate Immunity. Dev. Cell. 2015;34:45–57. doi: 10.1016/j.devcel.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson M.R., Hines K.M. Stromules: Probing formation and function. Plant Physiol. 2018;176:128–137. doi: 10.1104/pp.17.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Exposito-Rodriguez M., Laissue P.P., Yvon-Durocher G., Smirnoff N., Mullineaux P.M. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat. Commun. 2017;8:49. doi: 10.1038/s41467-017-00074-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asada K. Ascorbate peroxidase—A hydrogen peroxide-scavenging enzyme in plants. Physiol. Plant. 1992;85:235–241. doi: 10.1111/j.1399-3054.1992.tb04728.x. [DOI] [Google Scholar]
- 45.Dietz K.J. Thiol-based peroxidases and ascorbate peroxidases: Why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Mol. Cells. 2016;39:20–25. doi: 10.14348/molcells.2016.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002 doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 47.Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;7:405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 48.Shigeoka S., Ishikawa T., Tamoi M., Miyagawa Y., Takeda T., Yabuta Y., Yoshimura K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 2002;53:1305–1319. doi: 10.1093/jexbot/53.372.1305. [DOI] [PubMed] [Google Scholar]
- 49.Mano J., Ohno C., Domae Y., Asada K. Chloroplastic ascorbate peroxidase is the primary target of methylviologen-induced photooxidative stress in spinach leaves: Its relevance to monodehydroascorbate radical detected with in vivo ESR. Biochim. Biophys. Acta-Bioenerg. 2001;1504:275–287. doi: 10.1016/S0005-2728(00)00256-5. [DOI] [PubMed] [Google Scholar]
- 50.Karpinski S., Escobar C., Karpinska B., Creissen G., Mullineaux P.M. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in arabidopsis during excess light stress. Plant Cell. 1997;9:627–640. doi: 10.1105/tpc.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karpinski S., Reynolds H., Karpinska B., Wingsle G., Creissen G., Mullineaux P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284:654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- 52.Janku M., Luhová L., Petrivalský M. On the origin and fate of reactive oxygen species in plant cell compartments. Antioxidants. 2019;8:105. doi: 10.3390/antiox8040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sewelam N., Jaspert N., Van Der Kelen K., Tognetti V.B., Schmitz J., Frerigmann H., Stahl E., Zeier J., Van Breusegem F., Maurino V.G. Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol. Plant. 2014;7:1191–1210. doi: 10.1093/mp/ssu070. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi H., Chen Z., Du H., Liu Y., Klessig D.F. Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 1997;11:993–1005. doi: 10.1046/j.1365-313X.1997.11050993.x. [DOI] [PubMed] [Google Scholar]
- 55.Chaouch S., Queval G., Vanderauwera S., Mhamdi A., Vandorpe M., Langlois-Meurinne M., van Breusegem F., Saindrenan P., Noctor G. Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 2010;153:1692–1705. doi: 10.1104/pp.110.153957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H.K., Zabeau M., Van Montagu M., Inzé D., Van Breusegem F. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 2004;39:45–58. doi: 10.1111/j.1365-313X.2004.02105.x. [DOI] [PubMed] [Google Scholar]
- 57.Queval G., Issakidis-Bourguet E., Hoeberichts F.A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cel. Plant J. 2007;52:640–657. doi: 10.1111/j.1365-313X.2007.03263.x. [DOI] [PubMed] [Google Scholar]
- 58.Su T., Wang P., Li H., Zhao Y., Lu Y., Dai P., Ren T., Wang X., Li X., Shao Q., et al. The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 2018;60:591–607. doi: 10.1111/jipb.12649. [DOI] [PubMed] [Google Scholar]
- 59.Shumbe L., Chevalier A., Legeret B., Taconnat L., Monnet F., Havaux M. Singlet oxygen-induced cell death in arabidopsis under high-light stress is controlled by OXI1 kinase. Plant Physiol. 2016;170:1757–1771. doi: 10.1104/pp.15.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rentel M.C., Lecourieux D., Ouaked F., Usher S.L., Petersen L., Okamoto H., Knight H., Peck S.C., Grierson C.S., Hirt H., et al. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature. 2004;427:858–861. doi: 10.1038/nature02353. [DOI] [PubMed] [Google Scholar]
- 61.Beaugelin I., Chevalier A., D’Alessandro S., Ksas B., Novák O., Strnad M., Forzani C., Hirt H., Havaux M., Monnet F. OXI1 and DAD regulate light-induced cell death antagonistically through jasmonate and salicylate levels. Plant Physiol. 2019;180:1691–1708. doi: 10.1104/pp.19.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mateo A., Funck D., Mühlenbock P., Kular B., Mullineaux P.M., Karpinski S. Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 2006;57:1795–1807. doi: 10.1093/jxb/erj196. [DOI] [PubMed] [Google Scholar]
- 63.Keun P.L., Kim C., Landgraf F., Apel K. EXECUTER1-and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2007;104:10270–10275. doi: 10.1073/pnas.0702061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner D., Przybyla D., Op Den Camp R., Kim C., Landgraf F., Keun P.L., Würsch M., Laloi C., Nater M., Hideg E., et al. The genetic basis of singlet oxygen-induced stress response of Arabidopsis thaliana. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 65.Meskauskiene R., Nater M., Goslings D., Kessler F., Op den Camp R., Apel K. FLU: A negative regulator of chlorophyll biosynthesis in arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang S., Apel K., Kim C. Singlet oxygen-mediated and EXECUTER dependent signalling and acclimation of Arabidopsis thaliana exposed to light stress. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130227. doi: 10.1098/rstb.2013.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dogra V., Duan J., Lee K.P., Lv S., Liu R., Kim C. FtsH2-dependent proteolysis of EXECUTER1 is essential in mediating singlet oxygen-triggered retrograde signaling in Arabidopsis thaliana. Front. Plant Sci. 2017;8 doi: 10.3389/fpls.2017.01145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L., Kim C., Xu X., Piskurewicz U., Dogra V., Singh S., Mahler H., Apel K. Singlet oxygen-and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc. Natl. Acad. Sci. USA. 2016;113:E3792–E3800. doi: 10.1073/pnas.1603562113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dogra V., Li M., Singh S., Li M., Kim C. Oxidative post-translational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang L., Leister D., Guan L., Zheng Y., Schneider K., Lehmann M., Apel K., Kleine T. The Arabidopsis SAFEGUARD1 suppresses singlet oxygen-induced stress responses by protecting grana margins. Proc. Natl. Acad. Sci. USA. 2020;117:6918–6927. doi: 10.1073/pnas.1918640117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun X., Feng P., Xu X., Guo H., Ma J., Chi W., Lin R., Lu C., Zhang L. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2011;2:477. doi: 10.1038/ncomms1486. [DOI] [PubMed] [Google Scholar]
- 72.Isemer R., Krause K., Grabe N., Kitahata N., Asami T., Krupinska K. Plastid located WHIRLY1 enhances the responsiveness of Arabidopsis seedlings toward abscisic acid. Front. Plant Sci. 2012;3 doi: 10.3389/fpls.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316 doi: 10.1126/science.1140516. [DOI] [PubMed] [Google Scholar]
- 74.Page M.T., McCormac A.C., Smith A.G., Terry M.J. Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol. 2017;213:1168–1180. doi: 10.1111/nph.14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Susek R.E., Ausubel F.M., Chory J. Signal transduction mutants of arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 76.Nott A., Jung H.-S., Koussevitzky S., Chory J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- 77.Strand Å., Asami T., Alonso J., Ecker J.R., Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinix. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- 78.Mochizuki N., Tanaka R., Tanaka A., Masuda T., Nagatani A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2008;105:15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moulin M., McCormac A.C., Terry M.J., Smith A.G. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. USA. 2008;105:15178–15183. doi: 10.1073/pnas.0803054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Z.W., Yuan S., Feng H., Xu F., Cheng J., Shang J., Zhang D.W., Lin H.H. Transient accumulation of Mg-protoporphyrin IX regulates expression of PhANGs-New evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. J. Plant Physiol. 2011;168:714–721. doi: 10.1016/j.jplph.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 81.Woodson J.D., Perez-Ruiz J.M., Chory J. Heme synthesis by plastid ferrochelatase i regulates nuclear gene expression in plants. Curr. Biol. 2011;21:897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Page M.T., Garcia-Becerra T., Smith A.G., Terry M.J. Overexpression of chloroplast-targeted ferrochelatase 1 results in a genomes uncoupled chloroplast-to-nucleus retrograde signalling phenotype. Philos. Trans. R. Soc. B Biol. Sci. 2020;375:20190401. doi: 10.1098/rstb.2019.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tabrizi S.T., Sawicki A., Zhou S., Luo M., Willows R.D. GUN4-Protoporphyrin IX Is a singlet oxygen generator with consequences for plastid retrograde signaling. J. Biol. Chem. 2016;291:8978–8984. doi: 10.1074/jbc.C116.719989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Colombo M., Tadini L., Peracchio C., Ferrari R., Pesaresi P. GUN1, a jack-of-all-trades in chloroplast protein homeostasis and signaling. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tadini L., Pesaresi P., Kleine T., Rossi F., Guljamow A., Sommer F., Mühlhaus T., Schroda M., Masiero S., Pribil M., et al. Gun1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 2016;170:1817–1830. doi: 10.1104/pp.15.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Llamas E., Pulido P., Rodriguez-Concepcion M. Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genet. 2017;13:e1007022. doi: 10.1371/journal.pgen.1007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu G.Z., Chalvin C., Hoelscher M., Meyer E.H., Wu X.N., Bock R. Control of retrograde signaling by rapid turnover of Genomes Uncoupled1. Plant Physiol. 2018;176:2472–2495. doi: 10.1104/pp.18.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marino G., Naranjo B., Wang J., Penzler J.F., Kleine T., Leister D. Relationship of GUN1 to FUG1 in chloroplast protein homeostasis. Plant J. 2019;99:521–535. doi: 10.1111/tpj.14342. [DOI] [PubMed] [Google Scholar]
- 89.Wu G.Z., Meyer E.H., Richter A.S., Schuster M., Ling Q., Schöttler M.A., Walther D., Zoschke R., Grimm B., Jarvis R.P., et al. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants. 2019;5:525–538. doi: 10.1038/s41477-019-0415-y. [DOI] [PubMed] [Google Scholar]
- 90.Shimizu T., Kacprzak S.M., Mochizuki N., Nagatani A., Watanabe S., Shimada T., Tanaka K., Hayashi Y., Arai M., Leister D., et al. The retrograde signaling protein GUN1 regulates tetrapyrrole biosynthesis. Proc. Natl. Acad. Sci. USA. 2019;116:24900–24906. doi: 10.1073/pnas.1911251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA. 2012;109:5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shao N., Duan G.Y., Bock R. A mediator of singlet oxygen responses in chlamydomonas reinhardtii and arabidopsis identified by a luciferase-based genetic screen in algal cells. Plant Cell. 2013;25:4209–4226. doi: 10.1105/tpc.113.117390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shumbe L., D’Alessandro S., Shao N., Chevalier A., Ksas B., Bock R., Havaux M. Methylene Blue Sensitivity 1 (MBS1) is required for acclimation of Arabidopsis to singlet oxygen and acts downstream of β-cyclocitral. Plant Cell Environ. 2017;40:216–226. doi: 10.1111/pce.12856. [DOI] [PubMed] [Google Scholar]
- 94.Xiao Y., Savchenko T., Baidoo E.E.K., Chehab W.E., Hayden D.M., Tolstikov V., Corwin J.A., Kliebenstein D.J., Keasling J.D., Dehesh K. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell. 2012;149:1525–1535. doi: 10.1016/j.cell.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 95.Walley J., Xiao Y., Wang J.Z., Baidoo E.E., Keasling J.D., Shen Z., Briggs S.P., Dehesh K. Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2015;112:6212–6217. doi: 10.1073/pnas.1504828112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seemann M., Wegner P., Schünemann V., Bui B.T.S., Wolff M., Marquet A., Trautwein A.X., Rohmer M. Isoprenoid biosynthesis in chloroplasts via the methylerythritol phosphate pathway: The (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (GcpE) from Arabidopsis thaliana is a [4Fe-4S] protein. J. Biol. Inorg. Chem. 2005;10:131–137. doi: 10.1007/s00775-004-0619-z. [DOI] [PubMed] [Google Scholar]
- 97.Jiang J., Zeng L., Ke H., De La Cruz B., Dehesh K. Orthogonal regulation of phytochrome B abundance by stress-specific plastidial retrograde signaling metabolite. Nat. Commun. 2019;10:2904. doi: 10.1038/s41467-019-10867-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang J., Xiao Y., Chen H., Hu W., Zeng L., Ke H., Ditengou F.A., Devisetty U.K., Palme K., Maloof J.N., et al. Retrograde induction of phyB orchestrates ethylene-auxin hierarchy to regulate growth. Plant Physiol. 2020;183:1268–1280. doi: 10.1104/pp.20.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Estavillo G.M., Crisp P.A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., Giraud E., Whelan J., David P., Javot H., et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pornsiriwong W., Estavillo G.M., Chan K.X., Tee E.E., Ganguly D., Crisp P.A., Phua S.Y., Zhao C., Qiu J., Park J., et al. A chloroplast retrograde signal, 3’phosphoadenosine 5’-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife. 2017;6 doi: 10.7554/eLife.23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klein M., Papenbrock J. The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J. Exp. Bot. 2004;55:1809–1820. doi: 10.1093/jxb/erh183. [DOI] [PubMed] [Google Scholar]
- 102.Dichtl B. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wilson P.B., Estavillo G.M., Field K.J., Pornsiriwong W., Carroll A.J., Howell K.A., Woo N.S., Lake J.A., Smith S.M., Harvey Millar A., et al. The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 2009;58:299–317. doi: 10.1111/j.1365-313X.2008.03780.x. [DOI] [PubMed] [Google Scholar]
- 104.Ishiga Y., Watanabe M., Ishiga T., Tohge T., Matsuura T., Ikeda Y., Hoefgen R., Fernie A.R., Mysore K.S. The SAL-PAP chloroplast retrograde pathway contributes to plant immunity by regulating glucosinolate pathway and phytohormone signaling. Mol. Plant-Microbe Interact. 2017;30:829–841. doi: 10.1094/MPMI-03-17-0055-R. [DOI] [PubMed] [Google Scholar]
- 105.Zhao C., Wang Y., Chan K.X., Marchant D.B., Franks P.J., Randall D., Tee E.E., Chen G., Ramesh S., Phua S.Y., et al. Evolution of chloroplast retrograde signaling facilitates green plant adaptation to land. Proc. Natl. Acad. Sci. USA. 2019;116:5015–5020. doi: 10.1073/pnas.1812092116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang J., Vanneste S., Brewer P.B., Michniewicz M., Grones P., Kleine-Vehn J., Löfke C., Teichmann T., Bielach A., Cannoot B., et al. Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and pin polarity. Dev. Cell. 2011;20:855–866. doi: 10.1016/j.devcel.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 107.Kim B.H., Von Arnim A.G. FIERY1 regulates light-mediated repression of cell elongation and flowering time via its 3′(2′),5′-bisphosphate nucleotidase activity. Plant J. 2009;58:208–219. doi: 10.1111/j.1365-313X.2008.03770.x. [DOI] [PubMed] [Google Scholar]
- 108.Rodríguez V.M., Chételat A., Majcherczyk P., Farmer E.E. Chloroplastic phosphoadenosine phosphosulfate metabolism regulates basal levels of the prohormone jasmonic acid in Arabidopsis leaves. Plant Physiol. 2010;152:1335–1345. doi: 10.1104/pp.109.150474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Balparda M., Armas A.M., Estavillo G.M., Roschzttardtz H., Pagani M.A., Gomez-Casati D.F. The PAP/SAL1 retrograde signaling pathway is involved in iron homeostasis. Plant Mol. Biol. 2020;102:323–337. doi: 10.1007/s11103-019-00950-7. [DOI] [PubMed] [Google Scholar]
- 110.Pogson B.J., Woo N.S., Förster B., Small I.D. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13:602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 111.Jarvis P., López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013;14:787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- 112.Chan K.X., Phua S.Y., Crisp P., McQuinn R., Pogson B.J. Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu. Rev. Plant Biol. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854. [DOI] [PubMed] [Google Scholar]
- 113.Finkelstein R.R. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;5:765–771. doi: 10.1046/j.1365-313X.1994.5060765.x. [DOI] [Google Scholar]
- 114.Arenas-Huertero F., Arroyo A., Zhou L., Sheen J., León P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000;14:2085–2096. doi: 10.1101/gad.14.16.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huijser C., Kortstee A., Pego J., Weisbeek P., Wisman E., Smeekens S. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to abscisic acid Insensitive-4: Involvement of abscisic acid in sugar responses. Plant J. 2000;23:577–585. doi: 10.1046/j.1365-313x.2000.00822.x. [DOI] [PubMed] [Google Scholar]
- 116.Laby R.J., Kim D., Gibson S.I. The ram1 mutant of Arabidopsis exhibits severely decreased β-amylase activity. Plant Physiol. 2001;127:1798–1807. doi: 10.1104/pp.010723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rook F., Corke F., Card R., Munz G., Smith C., Bevan M.W. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001;26:421–433. doi: 10.1046/j.1365-313X.2001.2641043.x. [DOI] [PubMed] [Google Scholar]
- 118.Nakano T., Suzuki K., Fujimura T., Shinshi H. Genome-wide analysis of the ERF gene family in arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta-Gene Regul. Mech. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 120.Giraud E., van Aken O., Ho L.H.M., Whelan J. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of Alternative Oxidase1a. Plant Physiol. 2009;150:1286–1296. doi: 10.1104/pp.109.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Acevedo-Hernández G.J., León P., Herrera-Estrella L.R. Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 2005;43:506–519. doi: 10.1111/j.1365-313X.2005.02468.x. [DOI] [PubMed] [Google Scholar]
- 122.Woodson J.D., Perez-Ruiz J.M., Schmitz R.J., Ecker J.R., Chory J. Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 2013;73:1–13. doi: 10.1111/tpj.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guo H., Feng P., Chi W., Sun X., Xu X., Li Y., Ren D., Lu C., David Rochaix J., Leister D., et al. Plastid-nucleus communication involves calcium-modulated MAPK signalling. Nat. Commun. 2016;7:12173. doi: 10.1038/ncomms12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Z.W., Feng L.Y., Cheng J., Tang H., Xu F., Zhu F., Zhao Z.Y., Yuan M., Chen Y.E., Wang J.H., et al. The roles of two transcription factors, ABI4 and CBFA, in ABA and plastid signalling and stress responses. Plant Mol. Biol. 2013;83:445–458. doi: 10.1007/s11103-013-0102-8. [DOI] [PubMed] [Google Scholar]
- 125.Cottage A., Gray J.C. Timing the switch to phototrophic growth: A possible role of GUN1. Plant Signal. Behav. 2011;6:578–582. doi: 10.4161/psb.6.4.14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kerchev P.I., Pellny T.K., Vivancos P.D., Kiddle G., Hedden P., Driscoll S., Vanacker H., Verrier P., Hancock R.D., Foyer C.H. The transcription factor ABi4 Is required for the ascorbic acid-dependent regulation of growth and regulation of jasmonate-dependent defense signaling pathways in arabidopsis. Plant Cell. 2011;23:3319–3334. doi: 10.1105/tpc.111.090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martin G., Leivar P., Ludevid D., Tepperman J.M., Quail P.H., Monte E. Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat. Commun. 2016;7:11431. doi: 10.1038/ncomms11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kakizaki T., Matsumura H., Nakayama K., Che F.S., Terauchi R., Inaba T. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 2009;151:1339–1353. doi: 10.1104/pp.109.145987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kacprzak S.M., Mochizuki N., Naranjo B., Xu D., Leister D., Kleine T., Okamoto H., Terry M.J. Plastid-to-nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant Physiol. 2019;179:18–23. doi: 10.1104/pp.18.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Page M.T., Kacprzak S.M., Mochizuki N., Okamoto H., Smith A.G., Terry M.J. Seedlings lacking the PTM protein do not show a genomes uncoupled (Gun) mutant phenotype. Plant Physiol. 2017;174:21–26. doi: 10.1104/pp.16.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Terry M.J., Smith A.G. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front. Plant Sci. 2013;4:14. doi: 10.3389/fpls.2013.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bobik K., Burch-Smith T.M. Chloroplast signaling within, between and beyond cells. Front. Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De Dios Barajas-López J., Blanco N.E., Strand Å. Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim. Biophys. Acta-Mol. Cell Res. 2013;1833:425–437. doi: 10.1016/j.bbamcr.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 134.Feng P., Guo H., Chi W., Chai X., Sun X., Xu X., Ma J., Rochaix J.D., Leister D., Wang H., et al. Chloroplast retrograde signal regulates flowering. Proc. Natl. Acad. Sci. USA. 2016;113:10708–10713. doi: 10.1073/pnas.1521599113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu X., Chi W., Sun X., Feng P., Guo H., Li J., Lin R., Lu C., Wang H., Leister D., et al. Convergence of light and chloroplast signals for de-etiolation through ABI4-HY5 and COP1. Nat. Plants. 2016;2:16066. doi: 10.1038/nplants.2016.66. [DOI] [PubMed] [Google Scholar]
- 136.Waters M.T., Wang P., Korkaric M., Capper R.G., Saunders N.J., Langdale J.A. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ruckle M.E., DeMarco S.M., Larkin R.M. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell. 2007;19:3944–3960. doi: 10.1105/tpc.107.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Saini G., Meskauskiene R., Pijacka W., Roszak P., Sjögren L.L.E., Clarke A.K., Straus M., Apel K. Happy on norflurazon (hon) mutations implicate perturbance of plastid homeostasis with activating stress acclimatization and changing nuclear gene expression in norflurazon-treated seedlings. Plant J. 2011;65:690–702. doi: 10.1111/j.1365-313X.2010.04454.x. [DOI] [PubMed] [Google Scholar]
- 139.Sun X., Xu D., Liu Z., Kleine T., Leister D. Functional relationship between mTERF4 and GUN1 in retrograde signaling. J. Exp. Bot. 2016;67:3909–3924. doi: 10.1093/jxb/erv525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bode R., Ivanov A.G., Hüner N.P.A. Global transcriptome analyses provide evidence that chloroplast redox state contributes to intracellular as well as long-distance signalling in response to stress and acclimation in Arabidopsis. Photosynth. Res. 2016;128:287–312. doi: 10.1007/s11120-016-0245-y. [DOI] [PubMed] [Google Scholar]
- 141.Murata N., Takahashi S., Nishiyama Y., Allakhverdiev S.I. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta-Bioenerg. 2007;1767:414–421. doi: 10.1016/j.bbabio.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 142.Thanthrige N., Jain S., Bhowmik S.D., Ferguson B.J., Kabbage M., Mundree S., Williams B. Centrality of BAGs in Plant PCD, Stress Responses, and Host Defense. Trends Plant Sci. 2020 doi: 10.1016/j.tplants.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 143.Latrasse D., Benhamed M., Bergounioux C., Raynaud C., Delarue M. Plant programmed cell death from a chromatin point of view. J. Exp. Bot. 2016;67:5887–5900. doi: 10.1093/jxb/erw329. [DOI] [PubMed] [Google Scholar]
- 144.Czarnocka W., Van Der Kelen K., Willems P., Szechyńska-Hebda M., Shahnejat-Bushehri S., Balazadeh S., Rusaczonek A., Mueller-Roeber B., Van Breusegem F., Karpiński S. The dual role of LESION SIMULATING DISEASE 1 as a condition-dependent scaffold protein and transcription regulator. Plant Cell Environ. 2017;40:2644–2662. doi: 10.1111/pce.12994. [DOI] [PubMed] [Google Scholar]
- 145.Dietrich R.A., Delaney T.P., Uknes S.J., Ward E.R., Ryals J.A., Dangl J.L. Arabidopsis mutants simulating disease resistance response. Cell. 1994;77:565–577. doi: 10.1016/0092-8674(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 146.Dietrich R.A., Richberg M.H., Schmidt R., Dean C., Dangl J.L. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell. 1997;88:685–694. doi: 10.1016/S0092-8674(00)81911-X. [DOI] [PubMed] [Google Scholar]
- 147.Mateo A., Mühlenbock P., Rustérucci C., Chang C.C.C., Miszalski Z., Karpinska B., Parker J.E., Mullineaux P.M., Karpinski S. LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 2004;136:2818–2830. doi: 10.1104/pp.104.043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Karpiński S., Szechyńska-Hebda M., Wituszyńska W., Burdiak P. Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 2013;36:736–744. doi: 10.1111/pce.12018. [DOI] [PubMed] [Google Scholar]
- 149.Woodson J.D., Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Raghavendra A.S., Padmasree K. Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 2003;8:546–553. doi: 10.1016/j.tplants.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 151.Van Lis R., Atteia A. Control of mitochondrial function via photosynthetic redox signals. Photosynth. Res. 2004;79:133–148. doi: 10.1023/B:PRES.0000015409.14871.68. [DOI] [PubMed] [Google Scholar]
- 152.Noguchi K., Yoshida K. Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion. 2008;8:87–99. doi: 10.1016/j.mito.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 153.De Clercq I., Vermeirssen V., Van Aken O., Vandepoele K., Murcha M.W., Law S.R., Inzé A., Ng S., Ivanova A., Rombaut D., et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell. 2013;25:3472–3490. doi: 10.1105/tpc.113.117168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yoshida K., Terashima I., Noguchi K. Distinct roles of the cytochrome pathway and alternative oxidase in leaf photosynthesis. Plant Cell Physiol. 2006;47:22–31. doi: 10.1093/pcp/pci219. [DOI] [PubMed] [Google Scholar]
- 155.Yoshida K., Terashima I., Noguchi K. Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol. 2007;48:606–614. doi: 10.1093/pcp/pcm033. [DOI] [PubMed] [Google Scholar]
- 156.Yoshida K., Noguchi K. Differential gene expression profiles of the mitochondrial respiratory components in illuminated arabidopsis leaves. Plant Cell Physiol. 2009;50:1449–1462. doi: 10.1093/pcp/pcp090. [DOI] [PubMed] [Google Scholar]
- 157.Van Aken O., Zhang B., Law S., Narsai R., Whelan J. AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol. 2013;162:254–271. doi: 10.1104/pp.113.215996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ivanova A., Law S.R., Narsai R., Duncan O., Lee J.H., Zhang B., Van Aken O., Radomiljac J.D., van der Merwe M., Yi K.K., et al. A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates alternative oxidase1a expression in arabidopsis. Plant Physiol. 2014;165:1233–1254. doi: 10.1104/pp.114.237495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ho L.H.M., Giraud E., Uggalla V., Lister R., Clifton R., Glen A., Thirkettle-Watts D., Van Aken O., Whelan J. Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in arabidopsis. Plant Physiol. 2008;147:1858–1873. doi: 10.1104/pp.108.121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Norman C., Howell K.A., Millar A.H., Whelan J.M., Day D.A. Salicylic Acid Is an Uncoupler and Inhibitor of Mitochondrial Electron Transport. Plant Physiol. 2004;134:492–501. doi: 10.1104/pp.103.031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liao Y., Tian M., Zhang H., Li X., Wang Y., Xia X., Zhou J., Zhou Y., Yu J., Shi K., et al. Salicylic acid binding of mitochondrial alpha-ketoglutarate dehydrogenase E2 affects mitochondrial oxidative phosphorylation and electron transport chain components and plays a role in basal defense against tobacco mosaic virus in tomato. New Phytol. 2015;205:1296–1307. doi: 10.1111/nph.13137. [DOI] [PubMed] [Google Scholar]
- 162.Duan J., Lee K.P., Dogra V., Zhang S., Liu K., Caceres-Moreno C., Lv S., Xing W., Kato Y., Sakamoto W., et al. Impaired psii proteostasis promotes retrograde signaling via salicylic acid1. Plant Physiol. 2019;180:2182–2197. doi: 10.1104/pp.19.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ng S., Giraud E., Duncan O., Law S.R., Wang Y., Xu L., Narsai R., Carrie C., Walker H., Day D.A., et al. Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J. Biol. Chem. 2013;288:3449–3459. doi: 10.1074/jbc.M112.416727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Blanco N.E., Guinea-Díaz M., Whelan J., Strand Å. Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130231. doi: 10.1098/rstb.2013.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Baena-González E., Rolland F., Thevelein J.M., Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 166.Baena-González E., Sheen J. Convergent energy and stress signaling. Trends Plant Sci. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang Y., Li L., Qu L.J. Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 2016;58:106–118. doi: 10.1111/jipb.12377. [DOI] [PubMed] [Google Scholar]
- 168.Van Aken O., Whelan J. Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsi. Front. Plant Sci. 2012;3 doi: 10.3389/fpls.2012.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kmiecik P., Leonardelli M., Teige M. Novel connections in plant organellar signalling link different stress responses and signalling pathways. J. Exp. Bot. 2016;67:3793–3807. doi: 10.1093/jxb/erw136. [DOI] [PubMed] [Google Scholar]
- 170.Choudhury F.K., Rivero R.M., Blumwald E., Mittler R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017;90:856–867. doi: 10.1111/tpj.13299. [DOI] [PubMed] [Google Scholar]
- 171.Liu J.X., Howell S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016;211:418–428. doi: 10.1111/nph.13915. [DOI] [PubMed] [Google Scholar]
- 172.Van Aken O., Pogson B.J. Convergence of mitochondrial and chloroplastic ANAC017/PAP-dependent retrograde signalling pathways and suppression of programmed cell death. Cell Death Differ. 2017;24:955–960. doi: 10.1038/cdd.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ng S., Ivanova A., Duncan O., Law S.R., Van Aken O., De Clercq I., Wang Y., Carrie C., Xu L., Kmiec B., et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell. 2013;25:3450–3471. doi: 10.1105/tpc.113.113985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Overmyer K., Tuominen H., Kettunen R., Betz C., Langebartels C., Sandermann H., Kangasjarvi J. Ozone-Sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell. 2000;12:1849. doi: 10.1105/tpc.12.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fujibe T., Saji H., Arakawa K., Yabe N., Takeuchi Y., Yamamoto K.T. A Methyl Viologen-Resistant Mutant of Arabidopsis, Which Is Allelic to Ozone-Sensitive rcd1, Is Tolerant to Supplemental Ultraviolet-B Irradiation. Plant Physiol. 2004;134:275–285. doi: 10.1104/pp.103.033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jaspers P., Blomster T., Brosché M., Salojärvi J., Ahlfors R., Vainonen J.P., Reddy R.A., Immink R., Angenent G., Turck F., et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 2009;60:268–279. doi: 10.1111/j.1365-313X.2009.03951.x. [DOI] [PubMed] [Google Scholar]
- 177.Brosché M., Blomster T., Salojärvi J., Cui F., Sipari N., Leppälä J., Lamminmäki A., Tomai G., Narayanasamy S., Reddy R.A., et al. Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by RADICAL-INDUCED CELL DEATH1. PLoS Genet. 2014;10:e1004112. doi: 10.1371/journal.pgen.1004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Heiber I., Ströher E., Raatz B., Busse I., Kahmann U., Bevan M.W., Dietz K.J., Baier M. The redox imbalanced mutants of Arabidopsis differentiate signaling pathways for redox regulation of chloroplast antioxidant enzymes. Plant Physiol. 2007;143:1774–1788. doi: 10.1104/pp.106.093328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hiltscher H., Rudnik R., Shaikhali J., Heiber I., Mellenthin M., Meirelles Duarte I., Schuster G., Kahmann U., Baier M. The radical induced cell death protein 1 (RCD1) supports transcriptional activation of genes for chloroplast antioxidant enzymes. Front. Plant Sci. 2014;5 doi: 10.3389/fpls.2014.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vainonen J.P., Jaspers P., Wrzaczek M., Lamminmäki A., Reddy R.A., Vaahtera L., Brosché M., Kangasjärvi J. RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 2012;442:573–581. doi: 10.1042/BJ20111739. [DOI] [PubMed] [Google Scholar]
- 181.Inzé A., Vanderauwera S., Hoeberichts F.A., Vandorpe M., van Gaever T., van Breusegem F. A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 2012;35:308–320. doi: 10.1111/j.1365-3040.2011.02323.x. [DOI] [PubMed] [Google Scholar]
- 182.Shapiguzov A., Vainonen J.P., Hunter K., Tossavainen H., Tiwari A., Järvi S., Hellman M., Aarabi F., Alseekh S., Wybouw B., et al. Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. eLife. 2019;8 doi: 10.7554/eLife.43284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.D’alessandro S., Ksas B., Havaux M. Decoding β-cyclocitral-mediated retrograde signaling reveals the role of a detoxification response in plant tolerance to photooxidative stress. Plant Cell. 2018;30:2495–2511. doi: 10.1105/tpc.18.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.South P.F., Cavanagh A.P., Liu H.W., Ort D.R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science. 2019;363:eaat9077. doi: 10.1126/science.aat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kromdijk J., Głowacka K., Leonelli L., Gabilly S.T., Iwai M., Niyogi K.K., Long S.P. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science. 2016;354:857–861. doi: 10.1126/science.aai8878. [DOI] [PubMed] [Google Scholar]
- 186.Głowacka K., Kromdijk J., Kucera K., Xie J., Cavanagh A.P., Leonelli L., Leakey A.D.B., Ort D.R., Niyogi K.K., Long S.P. Photosystem II Subunit S overexpression increases the efficiency of water use in a field-grown crop. Nat. Commun. 2018;9:1–9. doi: 10.1038/s41467-018-03231-x. [DOI] [PMC free article] [PubMed] [Google Scholar]