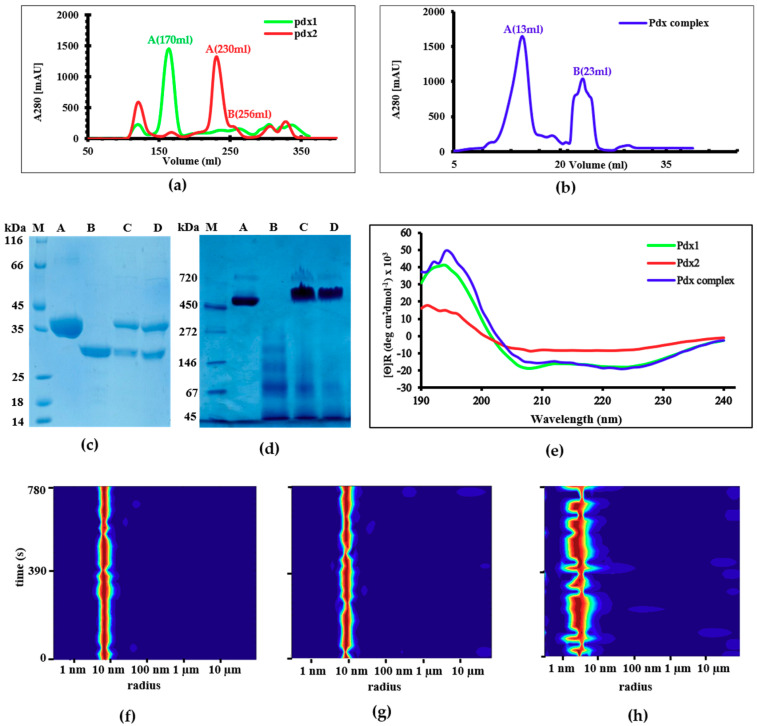
Figure 1.
Size-exclusion chromatography (SEC) elution profile of (a) Pdx1 and Pdx2 using a Superdex 200 26/60 column and (b) the Pdx complex using a Superose-6-increase 10/300 column; (c) SDS-PAGE of the purified samples: lane A corresponds to Pdx1, lane B to Pdx2, lane C to the Pdx1–Pdx2 complex before SEC, and D shows the SEC-purified Pdx complex; (d) Native PAGE showing the unreduced form of the Pdx proteins: lane A: Pdx1 dodecamer; lane B: Pdx2 in different oligomeric states; lane C: a Pdx1 and Pdx2 sample mixed in 1:1 molar ratio applied to SEC; lane D: purified Pdx complex at approximately 700 kDa; (e) Circular Dichroism (CD) spectroscopy results showing secondary structure contents of Pdx1 (green), the complex (blue) with α-helices and β-sheets as predominant secondary structure elements, and Pdx2 (red) with α-helices, β-sheets, and some random coils. Experimental CD data obtained for PvPdx2 were compared with different secondary structure content predicting servers, i.e., SOMPA (α-helices 33, β-sheets 21, and coils 40), DSSP (α-helices 29, β-sheets 32, and coils 39), and Predict Protein (α-helices 29, β-sheets 27, and coils 44). Calculated data are consistent with the experimental data. Dynamic light scattering (DLS) analysis of (f) dodecameric Pdx1 (RH = 7.3 ± 0.9 nm), (g) monodispersed saturated Pdx1–Pdx2 complex (RH = 9.7 ± 0.2 nm), and (h) monomeric Pdx2 (RH = 2.8 ± 0.4 nm).