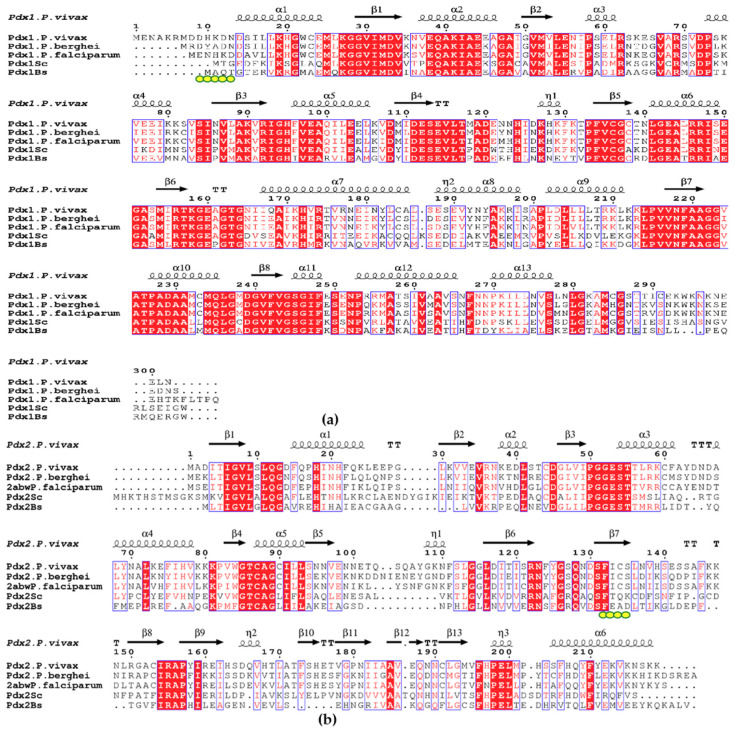
Figure 6.
(a) Alignment of Pdx1 amino acid sequences from P. vivax (A5K247) P. berghei (Q4Z0E8), P. falciparum (C6KT50), S. cerevisiae (Q03148), and B. subtilis (P37527); (b) Alignment of Pdx2 amino acid sequences from P. vivax (A0A1G4GXI4) P. berghei (Q4PJX5), P. falciparum (Q8IIK4), S. cerevisiae (Q03144), and B. subtilis (P37528). The active site residue of Pdx2 H-196 is mutated in PfPdx2 to N. The red boxes with white lettering show strict identity, red letters show similarity in the region between amino acid groups, and black letters show that regions are not conserved, with TT being a strict β turn and TTT a strict α turn. The Pdx1–Pdx2 contact surface involves many backbone interactions that are very much conserved in the three-dimensional structure but are not strictly conserved in the primary structure of Pdx2. Pivotal in the complex formation is the nonconserved region between β-5 and β-6 (η1) and the sequence region between β6 and β7 of the glutaminase. The yellow dots in Pdx1 mark the βN region, which interacts with the region (131–134) in the Pdx2 involved in β-completion and complex stabilization in bacterial complexes. This β-completion is not reported in plasmodial species [22], but the elongated N-terminus in PvPdx1 (which is absent in other plasmodial homologs) may have a role in complex stabilization. The sequence alignment was prepared by Espript3 [42].