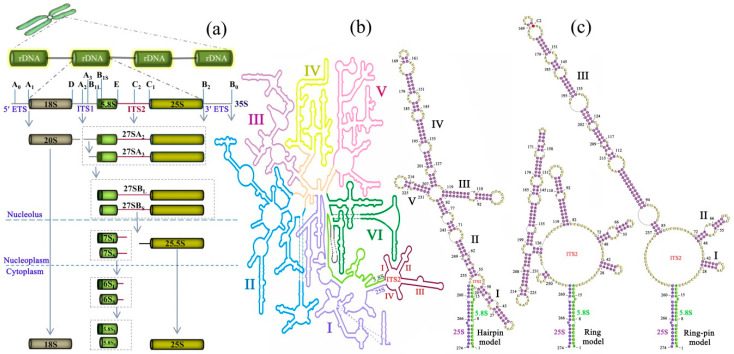
Figure 1.
Overview of the ITS2 (internal transcribed spacer 2) processing and folding in the ribosome biogenesis of the yeast S. cerevisiae. (a) ITS2 location and processing in pre-rRNA from genome to transcriptome. ITS2 and ITS1 are intercalated in the 18S–5.8S–25S tandem arrays, separating the elements of the pre-rRNA. The endonucleolytic cleavage sites are labelled A–E on the pre-rRNA. ITS2 region is highlighted in dark red in each processing state. (b) ITS2 location in the secondary structure of the 5.8S/25S pre-RNA. The ITS2 and 5.8S are displayed in dark red and bright green, respectively; the six typical domains of 25S are labelled I–VI in distinct colors. (c) The three proposed secondary structure models for ITS2. The C2 cleavage site is highlighted in red in stem III of the ring-pin model. The secondary structure scheme is taken and modified from S. cerevisiae LSU (http://apollo.chemistry.gatech.edu/RibosomeGallery/).