Abstract
Stress granules (SGs) represent important non-membrane cytoplasmic compartments, involved in cellular adaptation to various stressful conditions (e.g., hypoxia, nutrient deprivation, oxidative stress). These granules contain several scaffold proteins and RNA-binding proteins, which bind to mRNAs and keep them translationally silent while protecting them from harmful conditions. Although the role of SGs in cancer development is still poorly known and vary between cancer types, increasing evidence indicate that the expression and/or the activity of several key SGs components are deregulated in colorectal tumors but also in pre-neoplastic conditions (e.g., inflammatory bowel disease), thus suggesting a potential role in the onset of colorectal cancer (CRC). It is therefore believed that SGs formation importantly contributes to various steps of colorectal tumorigenesis but also in chemoresistance. As CRC is the third most frequent cancer and one of the leading causes of cancer mortality worldwide, development of new therapeutic targets is needed to offset the development of chemoresistance and formation of metastasis. Abolishing SGs assembly may therefore represent an appealing therapeutic strategy to re-sensitize colon cancer cells to anti-cancer chemotherapies. In this review, we summarize the current knowledge on SGs in colorectal cancer and the potential therapeutic strategies that could be employed to target them.
Keywords: Stress-Granules, Colorectal cancer, Adenylate-Uridylate-rich element-binding proteins, Post-transcriptional regulation, Oncogenes, Tumor suppressors
Core Tip: Colorectal cancer (CRC) represent the second cause of cancer mortality worldwide. Although changes in genetic landscape associated with CRC development have been identified, most frequent mutations are currently undruggable. The development of chemoresistance represent a major cause of CRC-associated mortality and identifying mechanisms allowing cancer cells to avoid these treatments may considerably improve clinical outcomes. Current findings indicate that cancers cells can preserve their expressed mRNAs in harmful conditions by storing them in small cytoplasmic granules, called Stress granules (SGs), where they are kept translationally silent. Targeting these SGs proteins may therefore represent a novel and efficient therapeutic approach.
INTRODUCTION
Colorectal cancer (CRC) represents the second cause of cancer mortality worldwide and the third most frequent cancer, with 1.8 million new cases and 881000 death in 2018[1]. CRC development results from a long term-deregulated process starting with the development of small adenomas, which evolve toward large adenomas and CRC. In most of the cases CRC develops sporadically (70%) and occurs in an aging population (> 50 years), whereas inherited genetic disorders such as Familial Adenomatous Polyposis or Lynch Syndrome are relatively rare and occurs at a younger age (before 50). Although the causes of sporadic CRC remain unclear, several risk factors have been identified, including inflammatory bowel disease (IBD) (e.g., Crohn’s disease and ulcerative colitis), obesity, diabetes, sedentary lifestyle, alcohol consumption, high fat-containing diet, and aging. Therefore, with the prevalence of obesity and diabetes worldwide, CRC incidence is expected to dramatically increase in the future, making this cancer a major public health concern and a growing economic burden. CRC is mostly treated by surgery, chemotherapy (e.g., FOLFOX: Folinic acid, 5-fluorouracil, oxaliplatin) and targeted therapy. However, despite these therapeutic options, the average survival rate of colon cancer between 2009 and 2015 was 63% (all SEER stages combined) and only 14% for distant CRC (American Cancer Society: https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis staging/survival-rates.html). This high mortality rate is predominantly due to metastasis and the development of chemoresistance[2]. Therefore, greater efforts are needed in identifying and targeting the mechanisms involved in both these processes in order to improve patient outcomes.
The development of chemoresistance is a major feature of CRC-associated mortality. Several chemoresistance mechanisms have been identified, including the induction of pro-survival factors and downregulation of pro-apoptotic proteins, along with the induction of transporters or detoxifications enzymes (e.g., P-glycoprotein), which reduce the efficiency of chemotherapy. More recently, it has been proposed that cancer cells can adapt to stress conditions (e.g., oxidative stress, hypoxia, chemotherapy) by generating small cytoplasmic ribonucleoprotein (RNP) foci called stress granules (SGs), which protect expressed mRNAs from degradation. SGs represent membrane-less cytoplasmic compartments containing mRNAs stalled at translation initiation. The mechanism underlying their formation is complex and tightly regulated by several proteins, which interact with mRNAs. SGs formation is also reversible, but in cases of prolonged stress, mRNAs are degraded into other cytoplasmic foci called processing bodies (P-Bodies). In cancer cells, SGs importantly contribute to cancer cell survival but also to resistance to various anti-cancerous agents. Several SGs components are upregulated in cancer cells as compared to their normal cellular counterparts. Moreover, several anti-cancerous agents elicit SGs assembly in cancer cells. Recent efforts aiming at identifying the mRNA/protein content of SGs have uncovered key players in carcinogenesis (e.g., oncogenes or tumor suppressors). Finally, impairment of SGs formation can re-sensitize several cancer types to chemotherapy or other anti-cancer agents (e.g., sorafenib) and thus may represent an appealing approach in combination with current treatments (e.g., FOLFOX, FOLFIRI)[3]. In this review, we discuss the role of SGs in colon cancer but also in pre-cancerous conditions favoring its development (e.g., inflammatory bowel disease). Because this cellular process has not been extensively studied in the context of CRC, we also discuss the current gaps in knowledge of SGs biology in CRC cells. Finally, we discuss potential therapeutic approaches that could be used to impact SGs assembly in cancer cells.
COMPLEXITY OF STRESS GRANULES FORMATION AND CAR-CINOGENESIS
Basics of SGs assembly
SGs are non-membrane cytoplasmic compartments, composed of untranslated RNPs formed in stressful conditions. SGs exhibit liquid-like behavior allowing rapid exchanges of components (e.g., mRNAs and proteins) with the cytosol[4,5]. The formation of SGs is a dynamic and conserved process, triggered by various stress conditions (e.g., nutrients deprivation, osmotic shock, hypoxia, heat shock, ultraviolet irradiation, oxidative stress), but also various molecules (e.g., chemotherapy, endoplasmic reticulum stressors, translation/proteasome inhibitors). Proteomic-based approaches have identified many of proteins located within mammalian SGs (https://msgp.pt/; http://rnagranuledb.lunenfeld.ca/). To date, more than 400 proteins have been identified in stress granules, but their composition may vary between cell types and/or stimuli. Among them, about 50% are RNA-Binding Proteins, while the remaining proteins are presumably recruited through protein-protein interaction and are involved in various cellular processes (e.g., cell cycle progression, apoptosis) or SGs assembly regulation.
The mechanisms involved in SGs formation are still unclear and several models have been proposed. SGs assembly is a multi-step process starting with the phosphorylation of eIF2α, which prevents the formation of the eIF2/GTP/tRNAi initiation complex[6] and leads to the dissociation of mRNAs from polysomes. However, this step is not mandatory for SGs assembly, as other non-canonical eIF2-independent models of SGs formation have been described (e.g., change in the activity of the eIF4F complex, which is also involved in translation initiation)[7,8]. Currently two models of SGs formation have been proposed[9]. In the “core first” model, untranslated mRNAs are nucleated into oligomers through the binding of proteins (e.g., T Cell-Restricted Intracellular Antigen-1, TIA1, G3BP1) having a Prion-Like Domain or Intrinsically Disordered Domains, which provide scaffolds necessary for the recruitment of other proteins (primary aggregation). These domains consist of polar residues, which favor liquid-liquid phase separation (LLPS) through electrostatic interactions. Due to these biophysical properties, SGs have been qualified as “liquid droplets”[10]. Then, the growth of these oligomers, through the addition of other untranslated RNPs give rise to the SGs “cores”. This step is supported by the microtubule’s cytoskeleton and motor proteins (e.g., dyneins, kinesins), which bring additional RNPs to the SGs[11]. Finally, the heterotypic associations of SGs components (e.g., G3BP1/TIA1; Polyadenylate-binding protein 1, PABP1) promote the growth and fusion of the granules (coalescence) and the recruitment of a dynamic shell, leading to the formation of large macroscopically visible SGs. However, this model has been challenged by the “LLPS First” model, where the nucleation of RNP generate phase separated droplets connected by weak interactions in which core granules are formed.
The formation of SGs is a tightly regulated process with participation of several signaling pathways and post-translational modifications (e.g., phosphorylation, acetylation)[12] of SGs components that regulate SGs assembly. For instance, phosphorylation of G3BP1 on ser149 by casein kinase 2 (CK2)[13] impairs SGs assembly, while arginine methylation, or deacetylation of G3BP1 by PRMT1/5 (protein arginine methyltransferase) and HDAC6[12], respectively[14,15], promotes their formation. Several signaling pathways regulate SGs assembly, including the PI3K or the Stress-Activated Kinase (p38/MAPK) signaling, which enhance SGs formation by activating mTORC1 kinase in stress conditions[16]. These pathways are usually overactivated in many cancers, following mutations of their key regulators (e.g., AKT, PTEN), thus providing a favorable landscape for SGs formation. However, it is still unclear the role of mTORC1 in SGs assembly, as other studies have suggested an opposite mechanism where AMPK inhibits mTORC1 and induces SGs assembly[17]. These differences may originate from the different models and stimuli used to trigger SGs. Finally, SGs formation is a reversible process and the clearance of SGs can be mediated by: (1) Translation re-initiation (after stress dissipates); (2) Chaperone proteins; (3) Autophagy (also referred as “granulophagy”); (4) mRNA degradation in processing bodies (P-Bodies); and (5) Proteasome-dependent degradation of SGs proteins.
While SGs formation is an adaptive response to physiological conditions where transient mRNA storage can occur, the role of SGs in human diseases have been recently recognized[6,18]. Dysregulation of SGs in various pathologies including neurodegenerative, viral infections, vascular diseases, and cancers indicate that SGs to be linked to disease progression. The mechanisms underlying SGs dysregulation in disease are not fully understood, yet aberrant expression of SGs components and altered pathway activity regulating their assembly and clearance appear to be contributing factors to the development of disease-associated SGs formation.
SGs and carcinogenesis
Overexpression of SGs assembly-related proteins, along with impairment of proteins/processes involved in their clearance are the main causes of SGs formation in cancers cells. Overexpression of nucleating proteins (e.g., G3BP1, TIA1, TIA-1-related, TIAR) is sufficient to trigger SGs formation in absence of stress[19]. Moreover, the stressful conditions present within the tumor microenvironment (e.g., hypoxia, oxidative stress, nutrient deprivation, chronic inflammation)[20], as well as specific molecules present (prostaglandin J2 and A1), promote SGs assembly in cancer cells[19]. Other factors including oxidized-low density lipoprotein or high-fat diet are also contributing factors to SGs formation[21]. These data suggest that lifestyle and chronic inflammatory/metabolic diseases (e.g., diabetes, fatty liver diseases, ulcerative colitis), which represent major risk factors for cancer development (e.g., hepatocellular carcinoma, CRC), may considerably influence SGs formation. Several anti-cancer treatments such as sorafenib, bortezomib, 5-FU, Oxaliplatin[19], FCCP [Carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone][22] and radiotherapy[19] increase SGs assembly in cancer cells, which in turn renders them more resistant to these treatments. SGs are also implicated in controlling cancer-related processes including apoptosis and migration/invasion of cancer cells. Although the precise mechanisms are still unclear, the sequestration and inhibition of pro-apoptotic factors (e.g., TRAF2, RACK1) has been suggested[23]. Furthermore, several RNA-binding proteins (e.g., tristetraprolin, HuR) located in SGs regulate the stability and translation of cancer-related mRNA transcripts involved in various cancerous hallmarks, including cell proliferation (e.g., MYC)[24], angiogenesis (e.g., VEGFA)[25], inflammation (e.g., cyclooxygenase-2, COX-2)[26] and cell death (antiapoptotic protein: BCL2, MCL1)[27,28]. SGs also inhibits cellular senescence by sequestering plasminogen activator inhibitor-1 in fibroblasts[29]. As senescence represents an important barrier against carcinogenesis[30], these findings further support the oncogenic function of SGs. Finally, defective P-body formation is observed in several cancers (e.g., CRC)[6], and this together with the increased SGs assembly may act in concert to promote tumor progression.
The current methodologies to study SGs functions are mostly based on gain and loss of function analyses of SGs components, microscopy, and cell fractionation methods to isolate SGs. This latest methodology, coupled with transcriptomic and proteomic-based approaches, have identified both proteins and mRNA transcripts associated with these granules. This information is publicly available in several databases (e.g., https://msgp.pt/; http://rnagranuledb.lunenfeld.ca/) and interestingly several transcripts and proteins have been associated to various cancer-related processes, suggesting that the role of SGs in carcinogenesis is largely underestimated. While the majority of studies utilize cell-based in vitro approaches, in vivo mouse models with constitutive deletion of specific SGs factors (e.g., TIA1KO or G3BP1KO mice) have been generated[31,32]. Further efforts utilizing tissue-specific and inducible knockout approaches will further aid in understanding the role of SGs play in development and progression of specific tumor types.
ROLE OF STRESS GRANULES IN COLORECTAL CANCER
Although the role of SGs has been studied in various cancers, their functions in the development of CRC and inflammatory bowel disease remain to be characterized. While more than 400 proteins have been identified in mammalian SGs (e.g., https://msgp.pt/), only few of them have been involved in SGs assembly and disassembly are abundantly expressed in epithelial and goblet cells of the colon (single cell sequencing of large intestine: https://tabula-muris.ds.czbiohub.org/) and also in the other cell types (e.g., enteroendocrine cells). In addition, several proteins (e.g., RNA-binding proteins, pro-apoptotic factors, cell cycle-related proteins) involved in cancer-related processes in CRC are localized in SGs.
SG nucleators and CRC
SGs nucleators refer to the proteins that are directly involved in the early aggregation phase of SGs formation and their sole overexpression is sufficient for spontaneous SGs assembly, even in absence of stress. Conversely, knockdown of these proteins severely impairs SGs assembly. Moreover, alteration of their expression occurs in preneoplasic conditions, such as ulcerative colitis, thus suggesting that defects in their expression are early alterations fostering CRC development.
UBAP2L: A component of the ubiquitin-proteasome pathway containing a ubiquitin-associated (UBA) domain which binds to ubiquitin and multi-ubiquitin chains[33]. UBAP2L plays a key role in stress granules assembly even in stress-null con-ditions[34,35]. The Arg-Gly-Gly (RGG) motif of UBAP2L plays a key role in SGs assembly[35] by mediating the recruitment of other components (e.g., RNPs)[35]. Importantly, this domain can be methylated by the protein arginine methyltransferase PRMT1, which impairs SGs formation[35]. The Domain of Unknown Function motif of UBAP2L is also necessary to bind to G3BP1/2[35]. Recent findings have suggested thus that UBAP2L acts upstream of G3BP1/2 and can form SGs core independently of G3BP1/2[36]. The function of UBAP2L in cancers is poorly known but increasing evidence indicates that UBAP2L promotes progression of hepatocellular carcinoma[37], prostate cancer[38] and glioblastoma[39]. The expression of UBAP2L in CRC is currently unknown (Table 1) but its knockdown in colon cancer cells (i.e., HCT116 and RKO cells)[40] hinders cell cycle progression[40] and induces apoptosis through activation of BAD, BAX, and the cleavage of Caspase-3 and Poly(ADP-ribose) Polymerase[40]. Although these results indicate an oncogenic function of UBAP2L in CRC, they are currently no studies documenting its function in SGs in CRC.
Table 1.
Expression and prognostic value of stress granule-associated proteins in colorectal cancer
| Components | Role in SG | mRNA expression in tumors (GEPIA) | Expression in CRC patients (literature) | Overall survival (GEPIA) | Overall survival (Human Protein Atlas) |
| G3BP1 | Promotes SG assembly | Up | Unknown | No significant difference | Better prognosis with high expression |
| G3BP2 | Promotes SG assembly | Up | Unknown | No significant difference | Better prognosis with high expression |
| USP10 | Promotes SG assembly | Up (trend) | Down in 18.6% of patients[47] | No significant difference | Better prognosis with high expression |
| CAPRIN1 | Promotes SG assembly | Up | Unknown | No significant difference | Better prognosis with high expression |
| UBAP2L | Promotes SG assembly | No significant difference | Unknown | No significant difference | No significant difference |
| TIA1 | Promotes SG assembly | Down (trend) | sTIA1 (spliced variant) is Up[171] | Poor prognosis with high expression | Better prognosis with high expression |
| TIAL1 | Promotes SG assembly | Down (trend) | Unknown | Poor prognosis with high expression | No significant difference |
| DDX3 | Promotes SG assembly | No significant difference | Poor prognosis with high expression[57] | No significant difference | Better prognosis with high expression |
| PABP1 | Promotes SG assembly | Up | Unknown | NA | No significant difference |
| FMR1 | Promotes SG assembly | No significant difference | Unknown | No significant difference | Better prognosis with high expression |
| PDCD4 | Promotes SG assembly | Down (trend) | Down[172] | No significant difference | No significant difference |
| ATXN2 | Promotes SG assembly | No significant difference | Unknown | No significant difference | No significant difference |
| ANG | Promotes SG assembly | No significant difference | Up | No significant difference | No significant difference |
| ZFP36 | Promotes SG clearance and SG-P-Bodies fusion | Down | Down[26] | Poor prognosis with low expression (P = 0.16: trend) | No significant difference |
| ZFP36L1 | Promotes SG-P-Bodies fusion | Down | Unknown | No significant difference | No significant difference |
| ELAVL1 | mRNA stabilization | Up (trend) | Up[26] | No significant difference | Better prognosis with high expression |
| CUGBP2 | mRNA stabilisation | No significant difference | Down[173] | No significant difference | Better prognosis with high expression |
| MSI-1 | Promotes SG assembly | No significant difference | Up[54] | No significant difference | No significant difference |
| KHSRP | Unknown | No significant difference | Unknown | No significant difference | No significant difference |
| BAG3 | Promotes SG clearance | No significant difference | Up[174] | No significant difference | Poor prognosis with high expression |
| PRMT1 | Inhibition of SG formation | Up (trend) | (Poor prognosis with high expression[119] | Better prognosis with high expression (not significant: marked trend) | Better prognosis with high expression |
| PRMT5 | Inhibition of SG formation | Up (trend) | Up[175] | Better prognosis with high expression (not significant: marked trend | Better prognosis with high expression |
| HDAC6 | Promotes SG assembly | Down | Up[176] | Poor prognosis with high expression (not significant: marked trend | Poor prognosis with high expression |
| SIRT6 | Promotes SG assembly | No significant difference | Down[113] | No significant difference | No significant difference |
| EP300 | Inhibition of SG formation | No significant difference | Up[105] | No significant difference | No significant difference |
| JMJD6 | Promotes SG assembly | No significant difference | Up[121] | No significant difference but marked trend for a poor prognosis with high expression | No significant difference |
| CK2 | Inhibition of SG formation | Up (trend) | Up[177] | No significant difference | Poor prognosis with high expression |
| PRKAA1 (AMPK) | Promotes SG assembly | No significant difference | Up[126,127] | No significant difference | Better prognosis with high expression |
| MTOR | Unclear | No significant difference | mTORC1 Up[122] | No significant difference | No significant difference |
| TARDBP | Promotes SG assembly | No significant difference | Unknown | No significant difference | No significant difference |
| RBFOX2 | Regulation of cell cycle | Down | Up[101] | No significant difference | No significant difference |
| RACK1 | Regulation of apoptosis | Up | No significant difference | No significant difference | |
| ULK1 | Promotes SG disassembly | Down | Up[178] (Poor Prognosis) | No significant difference | Poor prognosis with high expression |
| ULK2 | Promotes SG disassembly | Down | Down[179] | No significant difference | No significant difference |
| VCP | Promotes SG disassembly | No significant difference | Up (poor prognosis with high expression)[180] | No significant difference | No significant difference |
The differential mRNA expression of stress granule proteins in colorectal cancer as compared to matched non-tumoral tissues were retrieved from the GEPIA database (http://gepia.cancer-pku.cn/detail; normal tissues: n = 349; tumors: n = 275) and compared with published studies. Survival analyses were retrieved from the GEPIA (cutoff-High: 80%; cutoff-Low: 20%) and the Human Protein Atlas Database (https://www.proteinatlas.org) using the best separation method between low and high expression of protein candidates. SG: Stress granule; NA: Not available.
Ras GTPase-activating protein-binding protein (G3BP): A family of RNA-binding proteins composed of three different members, G3BP1, GBP2a and G3BP2b. Through their interaction with the SH3 domain of RasGAP (Ras GTPase activating protein), these proteins promote Ras signaling[41]. G3BP proteins contain a RNA recognition motif (RRM), which allows for interaction with the 40S subunit of ribosomes and RGG domains involved in mRNA binding[42]. Among them, G3BP1 is strongly overexpressed in a variety of cancers especially colon cancer[43] and exert oncogenic functions by promoting cancer cell proliferation, and inhibiting apoptosis[44]. Accordingly, downregulation of G3BP1 in colon cancer cells leads to a decrease of Ras signaling and cell growth arrest[43]. Despite the lack of a PLD, G3BP1 is an important SGs nucleator, as its sole overexpression is sufficient to trigger SGs assembly[45]. Although the mechanism involved is still unclear, recent studies have indicated that protein partners including CAPRIN1 or USP10, which promote and inhibit SGs assembly, respectively[46]. In CRC, the role of CAPRIN1 is currently unknown but a loss of USP10 expression was reported in 18.6% of CRC tumors[47]. Importantly, this loss was associated to distal metastasis and lymphovascular invasion.
The mechanism involved in G3BP1 overexpression is currently unknown but the RNA-binding protein Y-box binding protein (YB-1), which is overexpressed in CRC and ulcerative-associated lesions[48], may be involved in the increase of G3BP1 translation as suggested in other cancers[49]. G3BP1 is also regulated by post-translational modifications. G3BP1 phosphorylation on Ser149 by CK2 was reported to inhibits SGs formation[13]. However, the role of this phosphorylation is still controversial, as an erratum reporting that ser149 phosphorylation was unchanged during stress, has been published[50]. Arginine methylation in the RGG domain by protein arginine methyltransferase inhibits SGs formation[14]. Interestingly, this methylation is promoted by the Wnt/β-catenin pathway in mouse embryonic F9 cells[15], suggesting that overactivation of this oncogenic pathway in CRC may account for the increased SGs assembly in colon cancer cells. Finally, acetylation of Lysine-376 (K376) in the RRM domain, impairs the RNA binding function of G3BP1 as well as its interaction with PABP1, USP10 or Caprin1[12]. Accordingly, an increased acetylation of K376 was observed during SGs disassembly. HDAC6 and the CBP/p300 acetylase directly control the acetylation status of G3BP1[12] and thus HDAC6 inhibition impairs SGs formation[51]. While these findings indicate that the role of G3BP1 in SGs assembly, its role in CRC remains to be better characterized. Moreover, the other members of the G3BP family are potentially important for SGs assembly in colon cancer cells. For instance, G3BP2 overexpression can also trigger SGs formation in absence of stress[41,44]. The role of G3BP2 in CRC is currently unknown but in silico analysis of its mRNA level in CRC patients indicates an upregulation of G3BP2 in tumors as compared to surrounding non-tumoral tissue (Table 1).
TIA1 and TIAR: TIA1 is an RNA-binding protein comprised of three RRMs necessary for the binding to AU-Rich Elements (AREs) within the 3’UTR of target mRNAs and a PLD in C-terminal, which promotes self-aggregation of the protein. In stress conditions (i.e., hypoxia, oxidative stress), TIA1 interacts with co-factors (e.g., TIAR) to promotes the sequestration of target transcripts into SGs and inhibits their translation. In CRC, TIA1 expression is reduced. The mechanisms involved in TIA1 silencing haven’t been fully depicted but current findings indicate that the overexpression of miR-19a in CRC tissues and cell lines directly reduces TIA1 expression[52]. In CRC, TIA1 acts as a tumor suppressor by binding to the 3’UTR of COX-2 mRNA, thereby inhibiting its translation[53]. This tumor suppressive function is further supported with better prognosis observed in patients expressing a high level of TIA1 (Table 1). Intriguingly, TIA1 also contributes to chemoresistance to 5-FU in CRC cells[54]. These data suggest that although TIA1 exerts a tumor suppressive function in CRC, its role in SGs assembly may paradoxically favor cancer cell survival.
DEAD-Box RNA helicase 3 (DDX3 also called CAP-Rf): A ubiquitously expressed protein having an ATPase and helicase activity involved in RNA metabolism (e.g., mRNA splicing, transcription). DDX3 inhibits translation by directly interacting with eIF4E and with the SGs component PABP1, as evidenced in HeLa cells[55], indicating that this protein is important for SGs assembly. The role of DDX3 in SGs assembly is independent of its ATPase and helicase activity and downregulation of DDX3 in HeLa cells leads to a reduction of SGs formation, a re-localization of PABP1 to the nucleus, and an increased susceptibility to death stimuli (i.e., osmotic stress induced by sorbitol)[55]. However, the role of DDX3 in CRC is unclear with studies reporting both oncogenic[56,57] and tumor suppressive functions[58]. High tumor DDX3 expression correlates with a reduced survival in CRC patients[57]. Moreover, DDX3 expression is upregulated in colon biopsies from patients with inflammatory bowel disease[59], which may provide a favorable landscape for SGs formation. Interestingly, the DDX3 inhibitor RK-33 decreases expression of MMP-1, -2, -3 and -10 in HCEC1CT and HCEC2CT human colonic epithelial cells[59]. Interestingly, differentiation of HT-29 colon cancer cells is associated to decreased of DDX3 levels, suggesting that SGs formation is also influenced by the differentiation status of cancer cells[59].
G-quadruplex DNA structures (G4DNA): Current models of SGs formation have primarily focused on protein components triggering SGs assembly (e.g., G3BP, UBAP2L). However, recent studies have also highlighted the role of G-quadruplex DNA structures in liquid-liquid phase separation upon oxidative-stress-induced DNA damage. G4DNAs represent quartets of guanine linked by hydrogen bonds and organized as a planar ring[60]. Treatment of melanoma cells with hydrogen peroxide induces DNA damage and the production of ssDNA (single strand DNA), which forms G4DNA structures. Once exported into the cytosol, G4DNA interacts with various RNA-binding proteins involved in SG assembly, including YB-1, TIA1, TIAR, DHX36, and those involved in the control of the mRNA stability and translation (e.g., HuR)[60]. Accordingly, transfection of G4DNA is sufficient to trigger SG assembly in melanoma cells even in absence of stress, establishing G4DNA structures as potent SG nucleators[60]. The role of G4DNA in SG assembly in CRC cells has not been studied yet. However, increasing evidence indicate that G4DNA promotes CRC development and their inhibition with specific ligands (e.g., Emicoron) can promote antitumor activities[61].
tRNA-Derived stress-induced RNAs (tiRNA): Several non-coding RNAs, including microRNAs, long-non-coding RNAs and transfer RNAs have been involved in the adaptation of cells to stress stimuli[62]. Among them, tiRNA represent a novel class of non-coding RNAs generated in stress conditions by cleavage of mature tRNAs in the anticodon loop by angiogenin[62]. tiRNAs contribute to SGs formation by interacting with YB-1[62]. Moreover, tiRNAs can form G-quadruplex structures, which impair translation initiation by sequestering eIF4F complex[62]. Although the role of tiRNA in carcinogenesis is an emerging field, angiogenin is strongly upregulated in CRC as compared to non-tumoral tissues and promotes cancer progression by generating tiRNAs (e.g., 5’-tiRNA-val)[63]. Therefore, the accumulation of tiRNA together with the overexpression of nucleating proteins (e.g., G3BP) in colon cancer cells, provides a notable mechanism for SG assembly in absence of stress.
RNA-binding proteins controlling the expression of key oncogenes/tumor suppressors
During SGs formation, the binding of several RNA-binding proteins to their mRNA targets importantly regulate their stability and/or translation. Among them, Adenylate-Uridylate-rich elements binding proteins (AUBPs) represent critical post-transcriptional regulators of gene expression, through their ability to bind to AREs within the 3’UTR of mRNA transcripts and promote their recruitment toward P-bodies or SGs. Aberrant ARE-dependent post-transcriptional regulation has been associated to a variety of cancers, including CRC, by favoring the overexpression of oncogenes (e.g., c-myc) and pro-inflammatory mediators (e.g., COX-2), and the silencing of tumor suppressors (e.g., p53).
Tristetraprolin (TTP): TTP (ZFP36) belongs to a family of Cys-Cys-Cys-His zinc finger proteins and is an immediate-early response gene, whose expression can be induced by diverse stimuli such as insulin[64,65], TGF-b[66,67], LPS[68] and TNFa[69]. TTP is the best-characterized AU-Rich Element binding protein (AUBP) involved in promoting ARE-mediated mRNA decay. This process occurs through TTP-dependent delivery of ARE-mRNAs to P-bodies and recruit mRNA decay enzymes involved in deadenylation, decapping, and exonucleolytic decay[66,70-72]. TTP is also localized in SGs under conditions of energy deprivation[22]. However, the presence of TTP in SGs appears to be context-dependent as in models of oxidative stress, TTP is excluded from SGs due to phosphorylation of TTP by MK2[22]. Current findings suggest that TTP is involved in the shuttling between SGs and P-bodies[22] and SG-P-bodies fusion[73], and thus can contribute to SG clearance. TTP is considered as a tumor suppressor due to its capacity to reduce the expression of key inflammatory cytokines and also control expression of several factors involved in CRC carcinogenesis (e.g., COX-2, VEGFα)[74-76]. Accordingly, TTP expression is strongly reduced in colorectal tumors[26,77] as well as in early adenomas and adenocarcinomas, suggesting that early reduction of TTP may promote the establishment of a neoplasic phenotype. However, the link between TTP loss and SG dynamics in colon cancer cells remains unexplored.
Butyrate response factor 1 (TIS11b, ERF-1, cMGI, Berg36, ZFP36L1): An RNA-binding protein encoded by the ZFP36L1 gene, which belongs to the ZFP36 family[78,79]. Similar to TTP, BRF1 contains a tandem zinc finger domain bearing a double zinc finger motif (Cys-Cys-Cys-His) and promotes the decay of various cancer-promoting transcripts (e.g., VEGFA, cIAP2) by targeting them to P-bodies[79]. BRF1 is also a SG component and its overexpression promotes SG and P-body fusion[73]. However, the role of BRF1 in CRC is limited with only one study showing that 17-beta-oestradiol induces BRF1 in COLO205 colon cancer cells[80]. Nevertheless, in silico analyses of its mRNA level in CRC patients (Table 1) indicate a significant reduction of its expression similar to TTP, which may account for the deregulated expression pattern of various oncogenic transcripts. This downregulation may also reduce SG-P-body fusion in cancer cells, warranting further investigation.
HuR: A ubiquitously expressed RNA-binding protein encoded by the ELAVL1 gene, which belongs to the “Embryonic-Lethal Abnormal Vision in Drosophila” (ELAV) family[81]. HuR possess two tandem RRMs, followed by a hinge region and a third RRM. The HuR nucleocytoplasmic shuttling domain within the hinge region is subjected to phosphorylation by various kinases, which regulate the nucleo-cytoplasmic shuttling of the protein[82]. In the cytoplasm, HuR binds and stabilizes mRNA transcripts bearing an AU-rich sequences within their 3’UTR, by competing or displacing destabilizing factors (e.g., microRNAs, TTP)[26]. Moreover, HuR can directly bind and sequester miRNAs (e.g., miR-16, miR-21), thereby preventing the downregulation of their mRNA targets[83,84]. In stress conditions, HuR accumulates in SGs and promotes stabilization of various oncogenic transcripts[85]. However, other studies have suggested that the formation of SGs is dispensable for mRNA stabilization[86]. HuR is overexpressed in CRC as compared to normal tissues and exerts an oncogenic function by stabilizing the mRNAs of cancer and inflammatory-promoting factors involved in cancer cells proliferation, survival, and migration[26]. Moreover, HuR expression is also increased in colonic epithelial cells from patients with inflammatory bowel disease[87], thus adding another early event potentially fostering CRC development.
CUGBP2 (CUG-Binding Protein ELAV-like family member 2): CUGBP2 is a member of the CUGBP-ETR-3-like factors family that is ubiquitously expressed. This protein contains two N-terminal RRMs and one C-terminal RRM. CUGBP2 is a SG-associated RNA-binding protein involved in stabilizing and impairing the translation of bound target mRNAs[88]. Its expression is strongly reduced in various cancers and in CRC, CUGBP2 downregulation is mediated by Prostaglandin-E2 and its overexpression promotes apoptosis and mitotic catastrophe induced by radiation in colon cancer cells[89]. Furthermore, CUGBP2 overexpression in HCT-116 cells triggers cell cycle arrest in G2/M and an induction of apoptosis due to a direct binding to the 3’UTR of Mcl-1 mRNA and an impairment of its translation[90].
Musashi-1 (Msi-1): An RNA-binding protein, which promotes mRNA stability and translation inhibition[91]. Msi-1 is overexpressed in a variety of cancers and contributes to the overexpression of oncogenes (e.g., oncotachykinin 1 in breast cancer)[92] or cancer-promoting factors[91]. Msi-1 is also overexpressed in CRC and its knockdown severely impairs tumor growth in vitro and in vivo[93,94]. Moreover, Msi-1 overexpression enhances the development of CD44 positive-colon cancer stem cells and promotes chemoresistance in cells treated with 5-FU, by enhancing SGs assembly[54].
K-homology splicing regulator protein (KSRP): An RNA-binding protein involved in mRNA stability, splicing, transcription, as well as microRNA biogenesis[95,96]. In CRC, KSRP acts as a tumor suppressor by promoting the mRNA decay of β-catenin and iNOS transcripts[97,98]. In stress conditions (e.g., oxidative stress), KSRP localizes in SG[99,100]. However, it is unclear the role of KSRP in SGs and whether this event occurs in colorectal cancer cells.
Other proteins involved in CRC development
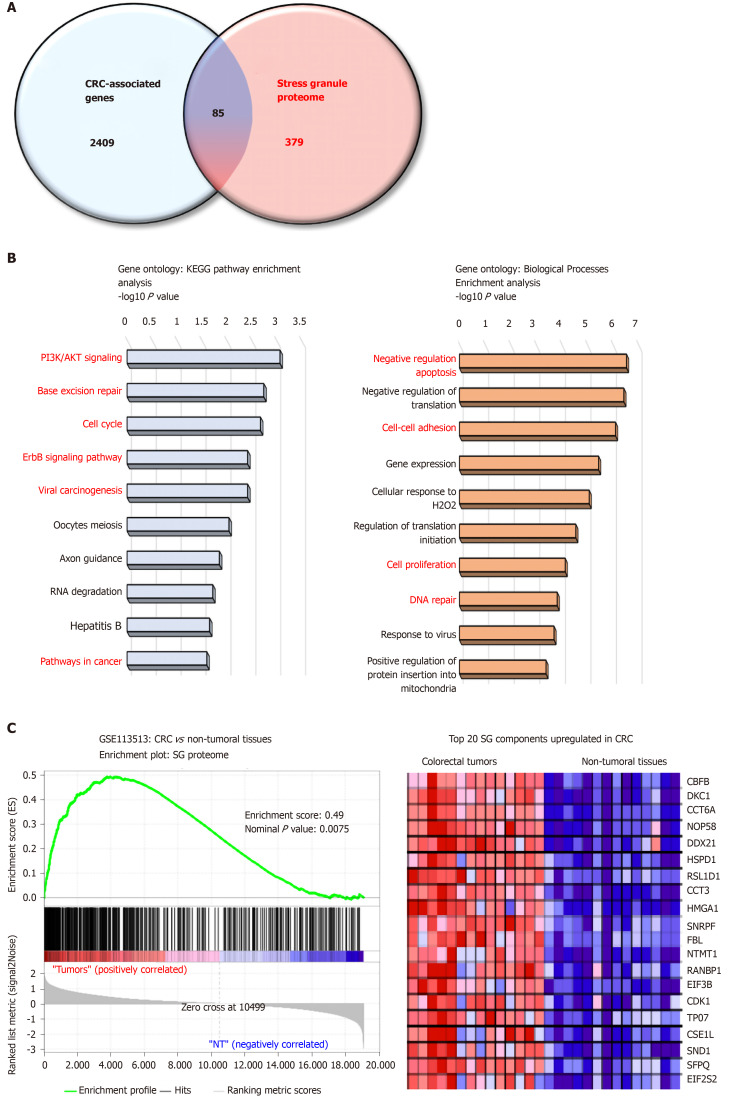
Proteome-based analysis has revealed more than 400 different proteins to be associated with SGs, and cross comparison with CRC-associated genes reveals 89 proteins (Figure 1A). Based on gene ontology analyses these proteins are involved in biological processes (negative regulation of apoptosis, cell adhesion, DNA repair) and pathways (e.g., PI3K, cell cycle) promoting colon carcinogenesis (Figure 1B and C). Moreover, gene set enrichment analysis indicates a significant enrichment of SG-associated genes in tumors as compared to non-tumor tissues (Figure 1B), with several oncogenes (e.g., CDK1, SND1, HSPD1) upregulated in CRC. Together, these data suggest that the SGs proteome represents an important “reservoir” of cancer-related factors, suggesting that the role of SGs in CRC is largely underestimated. Moreover, how various factors such as the genetic landscape (mutations), tumor etiology, lifestyle factors, and the gut microbiota influence the composition of the SGs proteome and tumor transcriptome is currently unknown. Therefore, it is likely that a different composition of SGs may differentially affect cancer-related processes based on intrinsic and external factors.
Figure 1.
The stress granule proteome contains several colorectal cancer-associated proteins. A: Venn diagram merging a list of colorectal cancer (CRC) -associated genes (retrieved from Metacore software) and the mammalian stress granule (SG) proteome from https://msgp.pt; B: Gene ontology analysis of SG proteome using KEGG pathway and biological processes analysis. Enrichment is represented with a-log10 P value. Processes and pathways in red are those involved in cancer development; C: Gene-Set Enrichment Analysis (version 3.0, Broad Institute, Cambridge, MA, United States) of the SG proteome on CRC patients (GSE113513). The top 20 genes upregulated in CRC patients as compared to non-tumoral tissues are represented in a heatmap. The enrichment score was calculated using the number of genes ranking at the top or the bottom of the gene list (permutation type: Phenotype; with 1000 permutations). The Signal2Noise was used for ranking genes. A nominal P value < 0.05 and an FDR < 0.2 were considered significant.
Several SG-associated proteins affect cancer hallmarks and pathways. For instance, under stress conditions the RBFOX2 (RBP fox-1 homolog 2) localizes in SGs and promotes cell cycle progression by decreasing RB1 protein expression in colon cancer cells[101]. Accordingly, RBFOX2 was found strongly upregulated in colon tumors, while RB1 was downregulated as compared to normal tissues. The pro-apoptotic factor RACK1 (Receptor for Activated C Kinase), which binds to and activates the stress responsive MTK1 MAPKKK[23], is sequestered into SGs in stress conditions and impairs its pro-apoptotic function. This effect has been also observed with chemotherapeutic agents such as 5-FU in HeLa cells, thus suggesting that RACK1 overexpression in CRC may also contribute to CRC chemoresistance[102]. In CRC, RACK1 is overexpressed and acts as a tumor promoter correlated with clinical outcomes[103]. Finally, PDCD4, another pro-apoptotic factor[104] is also localized in SGs[21] but it is currently unclear whether its sequestration in SGs impairs its pro-apoptotic function.
Post-translational modifications and SGs formation in CRC
Although several post-translational modifications of key proteins involved in SGs assembly have been identified, these alterations have yet to be studied in the CRC context. Nevertheless, it has been observed that the expression and activity of the proteins involved in modifying SGs factors can be altered in CRC.
Acetylation and deacetylation of SGs components: Acetylation of SG-associated proteins importantly regulate SGs assembly with several acetylases and deacetylases linked to this regulation. As previously discussed, acetylation of K376 of G3BP1 by the CBP/P300 acetylase is a key modification impeding SGs assembly by impairing its RNA binding function, as well as its interaction with PABP1, USP10 or Caprin1[12]. However, this link in CRC is unclear considering that the expression of CBP/P300 is upregulated in CRC[105,106]. Conversely, the histone deacetylase HDAC6 directly interacts and deacetylates G3BP1 and promotes SGs formation[12]. In agreement with these findings, SG disassembly is associated with increased acetylation of K376[12] and inactivation of HDAC6 catalytic domain impairs SGs formation in 293T cells[51]. The interaction of HDAC6 with G3BP1 is prevented by G3BP1 phosphorylation on Ser149[51]. Moreover, microtubules contribute to SG growth by supplying additional RNPs and other SG-associated proteins, are also subjected to acetylation modifications that markedly alter their dynamics. HDAC6 is also a microtubule-associated deacetylase[107] and its activity reduces tubulin-α acetylation on Lys40 in NIH3T3 cells and increases cell motility[107]. The activity of HDAC6 on microtubules and other motor proteins (e.g., dyneins) promotes SGs formation[51]. Taken together, these findings indicate that HDAC6 displays pleiotropic functions to promotes SGs assembly. While the role of HDAC6 in SGs assembly in CRC is currently unknown, its overexpression is observed in colorectal tumors compared to adjacent normal tissue and this may considerably favor SGs assembly[51]. Moreover, HDAC6 inhibitors can sensitize CRC cells to oxaliplatin[108].
SIRT6 is a NAD+-dependent deacetylase, which directly interacts with G3BP1. SIRT6 deficiency promotes G3BP1 phosphorylation at Ser149 and reduces SGs assembly[109]. Similar to HDAC6, the link between SIRT6 and SGs in CRC has not been established and discrepant observations have been reported regarding SIRT6 level in human CRC with studies reporting downregulation[110,111] or overexpression[112]. Nevertheless, SIRT6 overexpression correlates with a better prognosis in CRC patients and inhibits tumor progression[113,111].
Casein Kinase-2 (CK2): Phosphorylation of G3BP1 on Ser149 by CK2 impairs SGs formation in U2OS osteosarcoma cells[13]. This link has not been explored in CRC and the role of CK2 in colon cancer is currently unclear, with studies reporting both oncogenic and tumor suppressive functions[114,115]. CK2 expression and activity is increased in animal models of ulcerative colitis[116], suggesting that its altered expression represents an early alteration, potentially involved in colorectal carcinogenesis. In colon cancer cells, CK2 overexpression sensitizes cells to 5-FU[117,118] and promotes the degradation of several cancer-promoting transcripts by enhancing TTP function[115]. In contrast, CK2 enhances colon cancer cell viability by promoting COX-2 expression and PGE2 production[114].
Methylation of SG components: Protein Arginine Methyltransferase 1/5 (PRMT1/5) methylates several SGs components including G3BP1, G3BP2, FUS/LTS and UBAP2L. This methylation impairs the interaction of these SGs components (e.g., UBAP2L/G3BP1) and inhibits SGs assembly[35]. Paradoxically, various studies indicate an increase of SGs formation in colorectal cancer, PRMT1 and 5 are upregulated in tumors[119] (Table 1), suggesting that colon cancer cells adapt to circumvent this negative regulatory mechanism. A potential mechanism involves the histone arginine demethylase JMJD6 (Jumonji domain-containing 6), which promotes SGs formation by demethylating G3BP1[120]. In colon cancer JMJD6 is upregulated and exerts oncogenic functions (e.g., negative regulation of p53 signaling)[121]. Therefore, SG formation in colon cancer may depend on a fine-tuned equilibrium between PRMTs and JMJD6 activities.
AMPK/mTORC1 signaling: The role of AMPK and mTORC1 signaling in SGs formation is intriguing because in many models, SGs formation has been associated to a reduction of mTORC1 activity and/or activation of AMPK. However, in CRC mTORC1 activity is increased[122] and the link between mTORC1 inhibition and SGs assembly is likely to be cell type and context dependent[123]. In agreement with this, increased activity of mTORC1 by PI3K or p38 MAPK kinases has been associated to SGs formation in breast cancer cells[16]. These pathways are commonly overactive in CRC[124,125], and the downstream activation of mTORC1 may represent an important event triggering SGs assembly in colon cancer cells. The complexity mTORC1 signaling is further enhanced by AMPKα, which is an negative upstream regulator of mTORC1 signaling, in promoting SGs assembly[17] and is frequently upregulated in CRC[126,127]. Besides its regulatory function on SGs assembly, mTORC1 is also localized to SGs in stress conditions, thus impairing protein synthesis[128]. Once the stress signals dissipate, DYRK3 (Dual Specificity Tyrosine Phosphorylation Regulated Kinase 3) promotes the re-localization of mTORC1 to the cytosol to facilitate protein synthesis[128]. However, this regulatory mechanism remains to be demonstrated in the context of CRC.
SGs clearance in CRC
In addition of overexpression of nucleating proteins, alteration of SGs clearance contribute also to increased SGs in cancer cells. In this section, we discuss the various mechanisms involved in SGs dissolution and deregulated in colon cancer cells.
HspB8-HSP70-Bag3 complex: Several chaperones proteins are required for SGs clearance. In particular, the HspB8-HSP70 complex and co-chaperone protein Bag3 is involved in the “quality control” of SGs composition by preventing accumulation of misfolded ubiquitinated proteins in SGs[129]. Through an interaction with p62, an autophagy receptor accumulating in SGs[130], the complex targets misfolded proteins for degradation by autophagy[131]. Although they are currently no studies pertaining to HSPB8 in CRC, HSP70 and Bag3 are frequently overexpressed in CRC and contribute to cancer progression[132,133]. Although the HspB8-HSP70-Bag3 complex favors the maintenance of a normal SGs proteome function[129], this complex can also promote SGs disassembly in cases where stress persists. While this function remains to be better defined, some studies suggest that SGs clearance is mediated by autophagy[129].
Autophagy-dependent SGs clearance: Autophagy plays an important role in SGs clearance (also called “granulophagy”)[134]. Interestingly, autophagy and SGs are concomitantly increased in cancer cells[134] and have been recognized as important survival mechanisms in cancer cells, thus suggesting that these two processes act in concert to favor cancer cells survival. The role of autophagy in CRC development has been well characterized and like for many cancers, autophagic flux is strongly increased in CRC[135]. However, the link between autophagy and SGs clearance in CRC is currently unknown.
Valosin-containing protein (VCP/p97): An ATPase, which belongs to the AAA family (ATPases-associated with diverse cellular activities)[136]. Together with several cofactors, VCP interacts with ubiquitinated proteins and promotes their extraction from protein complexes for degradation[136]. This function is required during SGs disassembly[137]. In addition, VCP is essential for autophagosome maturation[138]. Accordingly, in several diseases (e.g., Inclusion body myopathy, Paget Disease), VCP mutations leads to a reduction of autophagy and an accumulation of SGs. VCP is also subjected to post-translational modifications, in particular phosphorylation by Unc-51-like kinases 1 and 2 (ULK1 and 2), which activates Vcp/p97 and causes SGs disassembly[137]. In CRC, ULK1 is overexpressed, while ULK2 is downregulated in tumors and the respective impact of these alterations on SG dynamics is unknown.
P-Bodies: Similar to SGs, P-bodies are also non-membrane RNA-protein complexes and their assembly is triggered by various stimuli including stress or inflammation[73]. In contrast to SGs, P-Bodies are mostly involved in mRNA decay and do not contain any translation initiation and elongation factors, but contain enzymes required for deadenylation (CAF-1, CCR4), decapping (DCP1/2), 5’ to 3’ degradation (XRN1) of mRNA transcripts[139,140]. mRNA degradation by P-bodies importantly contributes to SGs dissolution and disruption of P-body formation is likely to foster SGs accumulation[77]. Our full current understanding of the crosstalk between SGs and P-bodies is limited but appears to be a determinant of the fate of cancer-related transcripts. Some proteins like tristetraprolin (TTP) or BRF1, are localized in both compartments and can mediate the shuttling of mRNAs and promotes SG-P-body fusion[73,141]. Thus, the loss of TTP and BRF1 expression and activity in colon cancer cells may considerably alter SGs clearance. Moreover, the loss of TTP expression has been associated with a reduction of P-bodies in CRC cells[66,77].
SGS AS POTENTIAL BIOMARKERS AND THERAPEUTIC TARGETS
SGs as potential biomarkers
SGs importantly contribute to cancer cell survival to harmful conditions and represent an important barrier to chemotherapy. Assessing the expression of SGs nucleators in CRC biopsies may therefore represent a predictive approach to evaluate patient response to chemotherapy. However, the expression levels of several SGs components as well as their links with the clinical outcome are limited or unclear. Only one study has suggested that the presence of TIA-1 in tumor infiltrating lymphocytes represents a favorable survival predictive marker in colorectal cancer patients[142]. The use of public available database combining transcriptomic and survival analyses of CRC patients (GEPIA database: http://gepia.cancer-pku.cn/) can be useful to correlate the expression level of SGs components with clinical outcomes (Table 1). However, these correlations only consider respective mRNA levels in the analyses and these alterations do not necessarily translate at the protein level. As shown in Table 1, several discrepant findings between published observations and the transcriptomic data (e.g., USP10, PRMT1) can be observed. These discrepancies may also originate from heterogeneity between the patients and clinical samples evaluated. Finally, assessing the expression of individual SGs components may be insufficient considering that these proteins act in concert to promote SGs assembly. While using bioinformatic approaches to identify potential novel SG-based correlations are a notable starting point, validation efforts are still required to conclude the relevance of SGs as potential biomarkers for CRC.
SGs as therapeutic target
Inhibiting SGs formation may re-sensitize cancers cells to physiological death stimuli and anti-cancer agents (chemotherapy), as evidenced in various pre-clinical models[143]. Several strategies impairing SG assembly or SG-oncogenic activities have been developed and tested in various cancer cell types, which are discussed in the following section (Table 2).
Table 2.
Potential therapeutic approaches to impair stress granule function in cancer cells
| Strategies | Target | Models | Known impact on SGs | Anticancer effect on CRC | Clinical trials for CRC (ID) |
| Targeting proteins involved in SG assembly | |||||
| EGCG | G3BP1 | Lung cancer[146] | Reduction of SG assembly | Yes[181] | NCT02891538; NCT02321969; NCT01239095 |
| Resveratrol | G3BP1 | CRC[145] | Unknown | Yes[145] | NCT00433576; NCT00920803 |
| GAP161 peptide | G3BP1 | CRC[43] | Unknown | Yes[43] | None |
| RK-33 | DDX3 | CRC[182] | Unknown | Yes[182] | None |
| Targeting G4DNA/RNA structures | |||||
| EMICORON | G4DNA | CRC[147] | Unknown | Yes[147] | None |
| chANG | angiogenin | CRC[148] | Unknown | Yes[148] | None |
| (mAb), 26-2F | angiogenin | CRC[149] | Unknown | Yes[149] | None |
| Targeting AMPK/mTORC1 axis | |||||
| Compound C | AMPK | Yeast[151] CRC[152] | Impairs SG assembly in yeast[151] | Yes[152] | None |
| Rapamycin | mTORC1 | CRC[183] | Unknown | Yes[183] | NCT00409994; NCT03439462 |
| Everolimus | mTORC1 | Breast[16] | SG inhibition | Yes[184] | NCT01154335; NCT00419159; NCT01387880 |
| Temsirolimus | mTORC1 | CRC[185] | Unknown | Yes[185] | NCT00593060; NCT00827684; NCT01183663 |
| Targeting HDACS/SIRTs | |||||
| OSS_128167 | SIRT6 | Pancreas cancer[161] | Unknown | Unknown | No |
| A-452 | HDAC6 | CRC[158] | Unknown | Yes[158] | None |
| C1A | HDAC6 | CRC[157] | Unknown | Yes[157] | None |
| ACY-1215 | HDAC6 | CRC[108] | Unknown | Yes[108] | None |
| MPT0G612 | HDAC6 | CRC[159] | Unknown | Yes[159] | None |
| Targeting SGs-associated RNA-binding proteins controlling cancer-related factors | |||||
| MS-444 | HuR | CRC[166] | Unknown | Yes[166] | None |
| DHTS | HuR | CRC[186] | Unknown | Yes[186] | None |
| Resveratrol | RBFOX2 | CRC[101] | Unknown | Yes[101] | NCT00433576; NCT00920803 |
| Targeting microtubules | |||||
| Paclitaxel | Microtubules | Green monkey kidney fibroblasts (CV-1 cells)[153] | Promotes SG formation | Yes[187] | NCT00598247; NCT00024401; NCT00667641 |
| Vinblastine | Microtubules | Green monkey kidney fibroblasts (CV-1 cells)[153] | Prevents SG assembly | Yes[188] | None |
Several approaches can be used to efficiently reduce stress granule assembly and their oncogenic activities. This table provides some examples for each strategy. Some of them have been tested in colorectal cancers models and others have reached clinical trials (https://clinicaltrials.gov/). SG: Stress granule; CRC: Colorectal cancer.
Reducing the expression of SGs nucleators: As previously discussed, the inhibition or silencing of several proteins can efficiently prevent SGs assembly in cancer cells (e.g., G3BP1, UBAP2L). Therefore, developing therapeutic strategies to limit these SGs components specifically in cancer cells may represent a novel approach to reducing tumor growth and to re-sensitizing cells to chemotherapy. Although there are currently no studies assessing the therapeutic potential of inhibiting SGs in CRC, one approach using delivery of specific siRNAs (e.g., Aptamers) as a means to reduce expression of specific oncogenic targets has shown anti-tumor efficacy in CRC[144]. Such approaches could be also applied for microRNA delivery as a means to control the expression of SGs nucleators. Moreover, various small molecules have been shown to reduce the expression of SGs components in CRC cells, such as resveratrol or EGCG (Epigallocatechin-Gallate) for G3BP1[145,146]. Similarly, the peptide GAP161 can efficiently reduce G3BP1 activity and may represent a valuable tool to prevent SGs formation. This peptide markedly inhibits colon cancer cell proliferation by inducing apoptosis and sensitizing cells to cisplatin-induced apoptosis[43]. Furthermore, GAP161 reduces tumor growth in vivo, as evidenced in xenograft models. However, these antitumoral properties have been associated to an impairment of its interaction with RasGAP, so it is unclear whether SGs assembly is prevented in this model.
Targeting G4DNA/RNA structures: The importance of G4DNA/RNA structures in SGs assembly suggest them as novel therapeutic targets in various cancers. In agreement, the G-quadruplex ligand RHPS4 (3,11-difluoro-6,8,13-trimethyl-8Hquino) displays anti-tumor properties. However, this molecule also induces side effects such as cardiovascular alterations, suggesting caution regarding its clinical use. EMICORON, another G-quadruplex ligand displays also anti-tumor properties[147]. In colon cancer, EMICORON markedly reduces cancer progression[147] and potentiates chemotherapy in colon cancer cell lines[147].
A number of angiogenin inhibitors have been also developed and may reduce tiRNA accumulation in cancer cells. Among them, chANG, an antiangiogenin peptide, has been studied in colon cancer and shows antiangiogenic activity[148]. Moreover, a neutralizing monoclonal antibody to angiogenin prevents HT-29 colon cancer tumor progression in a xenograft model[149].
Targeting the AMPK/mTORC1 axis: SGs assembly is frequently associated to mTORC1 inhibition. Therefore, restoring normal mTORC1 activity has the potential to inhibit SGs assembly. However, increased activity of mTORC1 by PI3K or p38/MAPK kinases has been associated with SGs formation in MCF-7 breast cancer cells[150] and use of mTORC1 inhibitors may represent a potential therapeutic approach. Targeting upstream regulators of mTORC1, such as AMPK, which is a potent inhibitor of mTORC1 may also potentially impair SGs formation in cancer cells. The AMPK inhibitor Compound-C, efficiently prevents SGs assembly induced by a cold shock in yeast[151] and displays anti-cancer properties in colon cancer cells[152].
Targeting microtubules: The integrity of microtubules and motor proteins is required for RNP transport during SGs formation. Accordingly, microtubule destabilizing agents such as vinblastine or nocodazole can prevent SGs assembly, while drugs stabilizing them (e.g., paclitaxel) promote SGs formation[153]. However, vinblastine is currently not used clinically for CRC treatment due to its gastrointestinal toxicity[154]. Identifying new microtubule destabilizing agents with less side effects may potentially provide beneficial outcome to CRC patients. Several microtubule destabilizing agents have been developed and are currently used for the treatment of other cancers, such as eribulin for breast cancer[155,156].
HDAC and SIRT inhibitors: HDAC6 and SIRT6 activity promote SGs formation in cancer cells[51,109]. Several HDAC6 inhibitors have been developed such as A452, C1A, ACY-1215, MPT0G612, and have been shown to reduce CRC tumor growth and sensitize to cells to chemotherapeutic agents[108,157-159]. Targeting SIRT6 with specific inhibitors may also represent a potential approach to impair SGs assembly. However, only few SIRT6 inhibitors have been developed (e.g., OSS_128167)[160-162] and their effects on SGs assembly and CRC is currently unknown.
Targeting autophagy: The clearance of SGs is mediated by autophagy[163] and increasing autophagic flux in cancer cells may potentially lower the amount of SGs and re-sensitize cancer cells to chemotherapy. Alternatively, autophagy has been considered as an important survival mechanism of CRC cells and several molecules impairing autophagy have been implicated as novel therapeutics[135]. However, it remains to determine whether autophagy impairment can lead to an impairment of SGs clearance, which may potentially favor cancer cell survival and tumor recurrence.
Targeting SGs-associated RNA-binding proteins controlling cancer-related factors: Several RBPs are localized in SGs and control the translation/stability of various cancer-related transcripts (i.e., oncogenes, tumor suppressors). Targeting these proteins may represent an appealing approach to reduce the oncogenic properties of SGs in CRC. In that sense, several strategies aiming at inhibiting HuR expression and activity have been proposed[164]. Among them, the HuR inhibitor MS-444, a polyketides purified from microbial extracts, represents an interesting candidate due to its potent anti-cancerous properties in various cancers (e.g., colorectal cancer, pancreatic cancer, malignant glioma)[165,166]. MS-444 prevents HuR cytoplasmic export by inhibiting its homodimerization, thereby reducing the stability of its mRNA targets. Moreover, the anti-tumor properties of MS-444 was further observed in a mouse model of Familial Adenomatous Polyposis (i.e., APCMin mice)[87], thus showing the effectiveness of this molecule in vivo.
Finally, molecules preventing the sequestration of pro-apoptotic factors within SGs may also represent a potential approach to reduce cancer cell survival. For instance, resveratrol can prevent RBFOX2 localization in stress granules, thus inhibiting cancer cell proliferation[101].
CONCLUSION
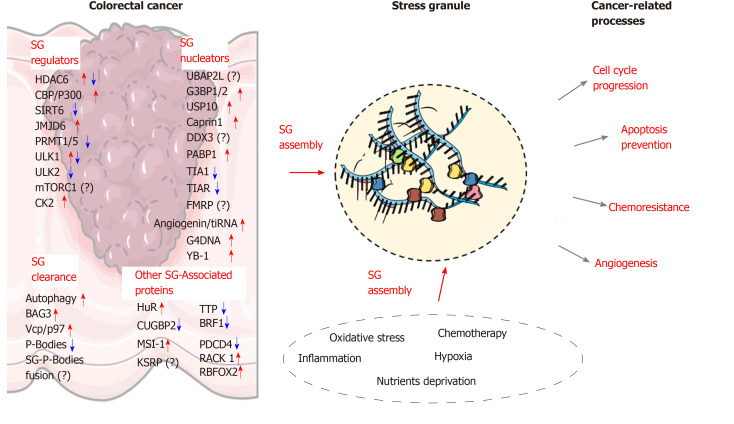
Due to the aging population and increased incidence of chronic bowel diseases, coupled with less than optimal lifestyle habits, an increased incidence in CRC cases is expected in the near future[167]. Further investigation into the molecular mechanisms associated with colon carcinogenesis is therefore needed in order to identify new targets for novel therapeutic approaches. Increasing evidence indicate that SGs are key players involved in CRC tumorigenesis and chemoresistance. Based on current findings, the assembly of SGs and their role in CRC development relies on multiple changes in the factors involved in SG nucleation and clearance (Figure 2). Early alterations of SGs components in preneoplastic conditions (e.g., IBD) may also allow altered cells to survive and accumulate further defects contributing to tumorigenesis. Moreover, the link between other chronic diseases such as diabetes and obesity, which are important risk factors for CRC development, represent new areas where SGs assembly needs to be clarified. Other potential contributing factors, such as gut microbiota dysbiosis should also be considered as a potential driver of SGs formation in cancer cells. Beside the proteins discussed in this review, several other SG components have been identified and have been recognized as regulators of SGs assembly in other cancer types, such as FMRP, ATX-2, the RNA helicase DDX3X[168], TDP-43[169], DYRK3[128], PDCD4[21] or FUS[170], and continued work will determine the function of these proteins in CRC. Other proteins, which are associated with SGs are also important regulators of cancerous processes (e.g., cell cycle progression, cancer cell migration and invasion). Although the role of these factors in SG biology is currently unclear, their storage in these granules may represent an important “reservoir”, favoring cancer cell survival in stress-related conditions. As CRC remains one of the deadliest cancers worldwide, employing strategies aimed at impairing SG assembly may re-sensitize cancer cells to chemotherapy and improve clinical outcomes. Such approaches may provide beneficial effects to CRC patients, along with other cancers where clinical options are limited and only a few therapeutic options exist. In this review, we discussed several strategies that could be employed to reduce SG formation in cancer cells. However, the efficiency of such approaches in colorectal cancer and SG assembly needs to be firmly established. Moreover, the potential side effects that could be associated with these strategies (e.g., the G-quadruplex ligand RHPS4 which induces cardiovascular side effects) need to be carefully evaluated using in vivo models. Moreover, the role of some regulators of SGs formation in CRC is still unclear (e.g., mTORC1, AMPK) and thus a better understanding of their function in SG formation in CRC is required prior to any therapeutic interventions.
Figure 2.
The molecular landscape underlying stress granules formation in colorectal cancer. Stress granules (SGs) assembly in colorectal cancer cells is associated with several alterations in the expression of proteins involved in SG nucleation or clearance. The stress-related conditions within the tumor microenvironment and various antitumor agents can further promote SGs assembly. Several SG-associated proteins (RNA-binding proteins or others) contribute to various cancer-related processes such as cell cycle progression, apoptosis inhibition, angiogenesis, and chemoresistance. Illustrations were retrieved from Servier Medical art (https://smart.servier.com/). SG: Stress granules.
Footnotes
Conflict-of-interest statement: The authors declare no conflicts of interest.
Manuscript source: Invited manuscript
Peer-review started: July 22, 2020
First decision: August 8, 2020
Article in press: September 3, 2020
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Switzerland
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kaplan MA S-Editor: Yan JP L-Editor: A P-Editor: Ma YJ
Contributor Information
Noémie Legrand, Department of Medicine, Faculty of Medicine, University of Geneva, Geneva CH-1211, Switzerland.
Dan A Dixon, Department of Molecular Biosciences, University of Kansas, Lawrence, Kansas, and University of Kansas Cancer Center, Lawrence, KS 66045, United States.
Cyril Sobolewski, Department of Cell Physiology and Metabolism, Faculty of Medicine, University of Geneva, Geneva CH-1211, Switzerland. cyril.sobolewski@unige.ch.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet. 2016;7:105–114. [PMC free article] [PubMed] [Google Scholar]
- 3.De Rosa M, Pace U, Rega D, Costabile V, Duraturo F, Izzo P, Delrio P. Genetics, diagnosis and management of colorectal cancer (Review) Oncol Rep. 2015;34:1087–1096. doi: 10.3892/or.2015.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849:861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panas MD, Ivanov P, Anderson P. Mechanistic insights into mammalian stress granule dynamics. J Cell Biol. 2016;215:313–323. doi: 10.1083/jcb.201609081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Protter DSW, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chernov KG, Barbet A, Hamon L, Ovchinnikov LP, Curmi PA, Pastré D. Role of microtubules in stress granule assembly: microtubule dynamical instability favors the formation of micrometric stress granules in cells. J Biol Chem. 2009;284:36569–36580. doi: 10.1074/jbc.M109.042879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gal J, Chen J, Na DY, Tichacek L, Barnett KR, Zhu H. The Acetylation of Lysine-376 of G3BP1 Regulates RNA Binding and Stress Granule Dynamics. Mol Cell Biol. 2019;39:e00052–19. doi: 10.1128/MCB.00052-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reineke LC, Tsai WC, Jain A, Kaelber JT, Jung SY, Lloyd RE. Casein Kinase 2 Is Linked to Stress Granule Dynamics through Phosphorylation of the Stress Granule Nucleating Protein G3BP1. Mol Cell Biol. 2017;37:e00596–16. doi: 10.1128/MCB.00596-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai WC, Gayatri S, Reineke LC, Sbardella G, Bedford MT, Lloyd RE. Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly. J Biol Chem. 2016;291:22671–22685. doi: 10.1074/jbc.M116.739573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bikkavilli RK, Malbon CC. Arginine methylation of G3BP1 in response to Wnt3a regulates β-catenin mRNA. J Cell Sci. 2011;124:2310–2320. doi: 10.1242/jcs.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heberle AM, Razquin Navas P, Langelaar-Makkinje M, Kasack K, Sadik A, Faessler E, Hahn U, Marx-Stoelting P, Opitz CA, Sers C, Heiland I, Schäuble S, Thedieck K. The PI3K and MAPK/p38 pathways control stress granule assembly in a hierarchical manner. Life Sci Alliance. 2019;2:e201800257. doi: 10.26508/lsa.201800257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahboubi H, Koromilas AE, Stochaj U. AMP Kinase Activation Alters Oxidant-Induced Stress Granule Assembly by Modulating Cell Signaling and Microtubule Organization. Mol Pharmacol. 2016;90:460–468. doi: 10.1124/mol.116.105494. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Liu B. Relationships between Stress Granules, Oxidative Stress, and Neurodegenerative Diseases. Oxid Med Cell Longev. 2017;2017:1809592. doi: 10.1155/2017/1809592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahboubi H, Stochaj U. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim Biophys Acta Mol Basis Dis. 2017;1863:884–895. doi: 10.1016/j.bbadis.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Herman AB, Silva Afonso M, Kelemen SE, Ray M, Vrakas CN, Burke AC, Scalia RG, Moore K, Autieri MV. Regulation of Stress Granule Formation by Inflammation, Vascular Injury, and Atherosclerosis. Arterioscler Thromb Vasc Biol. 2019;39:2014–2027. doi: 10.1161/ATVBAHA.119.313034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Y, Dong Z, Shang Q, Zhao H, Wang L, Guo C, Gao F, Zhang L, Wang Q. Pdcd4 Is Involved in the Formation of Stress Granule in Response to Oxidized Low-Density Lipoprotein or High-Fat Diet. PLoS One. 2016;11:e0159568. doi: 10.1371/journal.pone.0159568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 24.Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, Schaub FX, Sanduja S, Dixon DA, Blackshear PJ, Cleveland JL. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell. 2012;150:563–574. doi: 10.1016/j.cell.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurosu T, Ohga N, Hida Y, Maishi N, Akiyama K, Kakuguchi W, Kuroshima T, Kondo M, Akino T, Totsuka Y, Shindoh M, Higashino F, Hida K. HuR keeps an angiogenic switch on by stabilising mRNA of VEGF and COX-2 in tumour endothelium. Br J Cancer. 2011;104:819–829. doi: 10.1038/bjc.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishimaru D, Ramalingam S, Sengupta TK, Bandyopadhyay S, Dellis S, Tholanikunnel BG, Fernandes DJ, Spicer EK. Regulation of Bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol Cancer Res. 2009;7:1354–1366. doi: 10.1158/1541-7786.MCR-08-0476. [DOI] [PubMed] [Google Scholar]
- 28.Cui J, Placzek WJ. Post-Transcriptional Regulation of Anti-Apoptotic BCL2 Family Members. Int J Mol Sci. 2018;19:308. doi: 10.3390/ijms19010308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Omer A, Patel D, Lian XJ, Sadek J, Di Marco S, Pause A, Gorospe M, Gallouzi IE. Stress granules counteract senescence by sequestration of PAI-1. EMBO Rep. 2018;19:e44722. doi: 10.15252/embr.201744722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:S27–S31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- 31.Heck MV, Azizov M, Stehning T, Walter M, Kedersha N, Auburger G. Dysregulated expression of lipid storage and membrane dynamics factors in Tia1 knockout mouse nervous tissue. Neurogenetics. 2014;15:135–144. doi: 10.1007/s10048-014-0397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin S, Zekri L, Metz A, Maurice T, Chebli K, Vignes M, Tazi J. Deficiency of G3BP1, the stress granules assembly factor, results in abnormal synaptic plasticity and calcium homeostasis in neurons. J Neurochem. 2013;125:175–184. doi: 10.1111/jnc.12189. [DOI] [PubMed] [Google Scholar]
- 33.Madura K. The ubiquitin-associated (UBA) domain: on the path from prudence to prurience. Cell Cycle. 2002;1:235–244. [PubMed] [Google Scholar]
- 34.Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, Angers S, Morris Q, Fabian M, Côté JF, Gingras AC. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol Cell. 2018;69:517–532.e11. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 35.Huang C, Chen Y, Dai H, Zhang H, Xie M, Zhang H, Chen F, Kang X, Bai X, Chen Z. UBAP2L arginine methylation by PRMT1 modulates stress granule assembly. Cell Death Differ. 2020;27:227–241. doi: 10.1038/s41418-019-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cirillo L, Cieren A, Barbieri S, Khong A, Schwager F, Parker R, Gotta M. UBAP2L Forms Distinct Cores that Act in Nucleating Stress Granules Upstream of G3BP1. Curr Biol. 2020;30:698–707.e6. doi: 10.1016/j.cub.2019.12.020. [DOI] [PubMed] [Google Scholar]
- 37.Li Q, Wang W, Hu YC, Yin TT, He J. Knockdown of Ubiquitin Associated Protein 2-Like (UBAP2L) Inhibits Growth and Metastasis of Hepatocellular Carcinoma. Med Sci Monit. 2018;24:7109–7118. doi: 10.12659/MSM.912861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D, Huang Y. Knockdown of ubiquitin associated protein 2-like inhibits the growth and migration of prostate cancer cells. Oncol Rep. 2014;32:1578–1584. doi: 10.3892/or.2014.3360. [DOI] [PubMed] [Google Scholar]
- 39.Zhao B, Zong G, Xie Y, Li J, Wang H, Bian E. Downregulation of ubiquitin-associated protein 2-like with a short hairpin RNA inhibits human glioma cell growth in vitro. Int J Mol Med. 2015;36:1012–1018. doi: 10.3892/ijmm.2015.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chai R, Yu X, Tu S, Zheng B. Depletion of UBA protein 2-like protein inhibits growth and induces apoptosis of human colorectal carcinoma cells. Tumour Biol. 2016;37:13225–13235. doi: 10.1007/s13277-016-5159-y. [DOI] [PubMed] [Google Scholar]
- 41.French J, Stirling R, Walsh M, Kennedy HD. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem J. 2002;34:223–231. doi: 10.1023/a:1021737413055. [DOI] [PubMed] [Google Scholar]
- 42.Götte B, Panas MD, Hellström K, Liu L, Samreen B, Larsson O, Ahola T, McInerney GM. Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery. PLoS Pathog. 2019;15:e1007842. doi: 10.1371/journal.ppat.1007842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Zhang S, He H, Zhao W, Chen J, Shao RG. GAP161 targets and downregulates G3BP to suppress cell growth and potentiate cisplaitin-mediated cytotoxicity to colon carcinoma HCT116 cells. Cancer Sci. 2012;103:1848–1856. doi: 10.1111/j.1349-7006.2012.02361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuki H, Takahashi M, Higuchi M, Makokha GN, Oie M, Fujii M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells. 2013;18:135–146. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- 45.Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim K, Huh T, Park Y, Koo DH, Kim H, Hwang I, Choi CH, Yi JM, Chung JY. Prognostic significance of USP10 and p14ARF expression in patients with colorectal cancer. Pathol Res Pract. 2020;216:152988. doi: 10.1016/j.prp.2020.152988. [DOI] [PubMed] [Google Scholar]
- 48.Fogt F, Poremba C, Shibao K, Itoh H, Kohno K, Zimmerman RL, Görtz HG, Dockhorn-Dworniczak B, Urbanski SJ, Alsaigh N, Heinz D, Noffsinger AE, Shroyer KR. Expression of survivin, YB-1, and KI-67 in sporadic adenomas and dysplasia-associated lesions or masses in ulcerative colitis. Appl Immunohistochem Mol Morphol. 2001;9:143–149. doi: 10.1097/00129039-200106000-00007. [DOI] [PubMed] [Google Scholar]
- 49.Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, Grunewald TG, Zhang F, Ng T, Delattre O, Evdokimova V, Wang Y, Gleave M, Sorensen PH. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208:913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panas MD, Kedersha N, Schulte T, Branca RM, Ivanov P, Anderson P. Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J Cell Biol. 2019;218:2425–2432. doi: 10.1083/jcb.201801214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Liu R, Yang F, Cheng R, Chen X, Cui S, Gu Y, Sun W, You C, Liu Z, Sun F, Wang Y, Fu Z, Ye C, Zhang C, Li J, Chen X. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16:53. doi: 10.1186/s12943-017-0625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixon DA, Balch GC, Kedersha N, Anderson P, Zimmerman GA, Beauchamp RD, Prescott SM. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J Exp Med. 2003;198:475–481. doi: 10.1084/jem.20030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiou GY, Yang TW, Huang CC, Tang CY, Yen JY, Tsai MC, Chen HY, Fadhilah N, Lin CC, Jong YJ. Musashi-1 promotes a cancer stem cell lineage and chemoresistance in colorectal cancer cells. Sci Rep. 2017;7:2172. doi: 10.1038/s41598-017-02057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J. 2012;441:119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 56.Wu DW, Lin PL, Cheng YW, Huang CC, Wang L, Lee H. DDX3 enhances oncogenic KRAS-induced tumor invasion in colorectal cancer via the β-catenin/ZEB1 axis. Oncotarget. 2016;7:22687–22699. doi: 10.18632/oncotarget.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He TY, Wu DW, Lin PL, Wang L, Huang CC, Chou MC, Lee H. DDX3 promotes tumor invasion in colorectal cancer via the CK1ε/Dvl2 axis. Sci Rep. 2016;6:21483. doi: 10.1038/srep21483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bol GM, Xie M, Raman V. DDX3, a potential target for cancer treatment. Mol Cancer. 2015;14:188. doi: 10.1186/s12943-015-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tantravedi S, Vesuna F, Winnard PT, Jr, Van Voss MRH, Van Diest PJ, Raman V. Role of DDX3 in the pathogenesis of inflammatory bowel disease. Oncotarget. 2017;8:115280–115289. doi: 10.18632/oncotarget.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Byrd AK, Zybailov BL, Maddukuri L, Gao J, Marecki JC, Jaiswal M, Bell MR, Griffin WC, Reed MR, Chib S, Mackintosh SG, MacNicol AM, Baldini G, Eoff RL, Raney KD. Evidence That G-quadruplex DNA Accumulates in the Cytoplasm and Participates in Stress Granule Assembly in Response to Oxidative Stress. J Biol Chem. 2016;291:18041–18057. doi: 10.1074/jbc.M116.718478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Porru M, Artuso S, Salvati E, Bianco A, Franceschin M, Diodoro MG, Passeri D, Orlandi A, Savorani F, D'Incalci M, Biroccio A, Leonetti C. Targeting G-Quadruplex DNA Structures by EMICORON Has a Strong Antitumor Efficacy against Advanced Models of Human Colon Cancer. Mol Cancer Ther. 2015;14:2541–2551. doi: 10.1158/1535-7163.MCT-15-0253. [DOI] [PubMed] [Google Scholar]
- 62.Tao EW, Cheng WY, Li WL, Yu J, Gao QY. tiRNAs: A novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J Cell Physiol. 2020;235:683–690. doi: 10.1002/jcp.29057. [DOI] [PubMed] [Google Scholar]
- 63.Li S, Shi X, Chen M, Xu N, Sun D, Bai R, Chen H, Ding K, Sheng J, Xu Z. Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int J Cancer. 2019;145:1395–1407. doi: 10.1002/ijc.32245. [DOI] [PubMed] [Google Scholar]
- 64.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265:16556–16563. [PubMed] [Google Scholar]
- 65.Cao H, Urban JF, Jr, Anderson RA. Insulin increases tristetraprolin and decreases VEGF gene expression in mouse 3T3-L1 adipocytes. Obesity (Silver Spring) 2008;16:1208–1218. doi: 10.1038/oby.2008.65. [DOI] [PubMed] [Google Scholar]
- 66.Blanco FF, Sanduja S, Deane NG, Blackshear PJ, Dixon DA. Transforming growth factor β regulates P-body formation through induction of the mRNA decay factor tristetraprolin. Mol Cell Biol. 2014;34:180–195. doi: 10.1128/MCB.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogawa K, Chen F, Kim YJ, Chen Y. Transcriptional regulation of tristetraprolin by transforming growth factor-beta in human T cells. J Biol Chem. 2003;278:30373–30381. doi: 10.1074/jbc.M304856200. [DOI] [PubMed] [Google Scholar]
- 68.Chen YL, Jiang YW, Su YL, Lee SC, Chang MS, Chang CJ. Transcriptional regulation of tristetraprolin by NF-κB signaling in LPS-stimulated macrophages. Mol Biol Rep. 2013;40:2867–2877. doi: 10.1007/s11033-012-2302-8. [DOI] [PubMed] [Google Scholar]
- 69.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 70.Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 72.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee SR, Jin H, Kim WT, Kim WJ, Kim SZ, Leem SH, Kim SM. Tristetraprolin activation by resveratrol inhibits the proliferation and metastasis of colorectal cancer cells. Int J Oncol. 2018;53:1269–1278. doi: 10.3892/ijo.2018.4453. [DOI] [PubMed] [Google Scholar]
- 75.Cha HJ, Lee HH, Chae SW, Cho WJ, Kim YM, Choi HJ, Choi DH, Jung SW, Min YJ, Lee BJ, Park SE, Park JW. Tristetraprolin downregulates the expression of both VEGF and COX-2 in human colon cancer. Hepatogastroenterology. 2011;58:790–795. [PubMed] [Google Scholar]
- 76.Lee HH, Yang SS, Vo MT, Cho WJ, Lee BJ, Leem SH, Lee SH, Cha HJ, Park JW. Tristetraprolin down-regulates IL-23 expression in colon cancer cells. Mol Cells. 2013;36:571–576. doi: 10.1007/s10059-013-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sobolewski C, Sanduja S, Blanco FF, Hu L, Dixon DA. Histone Deacetylase Inhibitors Activate Tristetraprolin Expression through Induction of Early Growth Response Protein 1 (EGR1) in Colorectal Cancer Cells. Biomolecules. 2015;5:2035–2055. doi: 10.3390/biom5032035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 79.Sanduja S, Blanco FF, Dixon DA. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2011;2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qiu Y, Langman MJ, Eggo MC. Targets of 17beta-oestradiol-induced apoptosis in colon cancer cells: a mechanism for the protective effects of hormone replacement therapy? J Endocrinol. 2004;181:327–337. doi: 10.1677/joe.0.1810327. [DOI] [PubMed] [Google Scholar]
- 81.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017:8. doi: 10.1002/wrna.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Poria DK, Guha A, Nandi I, Ray PS. RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene. 2016;35:1703–1715. doi: 10.1038/onc.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol Cancer Res. 2012;10:167–180. doi: 10.1158/1541-7786.MCR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 86.Bley N, Lederer M, Pfalz B, Reinke C, Fuchs T, Glaß M, Möller B, Hüttelmaier S. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 2015;43:e26. doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lang M, Berry D, Passecker K, Mesteri I, Bhuju S, Ebner F, Sedlyarov V, Evstatiev R, Dammann K, Loy A, Kuzyk O, Kovarik P, Khare V, Beibel M, Roma G, Meisner-Kober N, Gasche C. HuR Small-Molecule Inhibitor Elicits Differential Effects in Adenomatosis Polyposis and Colorectal Carcinogenesis. Cancer Res. 2017;77:2424–2438. doi: 10.1158/0008-5472.CAN-15-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, Fulzele A, Wozniak JM, Gonzalez DJ, Kankel MW, Gao FB, Bennett EJ, Lécuyer E, Yeo GW. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell. 2018;172:590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Natarajan G, Ramalingam S, Ramachandran I, May R, Queimado L, Houchen CW, Anant S. CUGBP2 downregulation by prostaglandin E2 protects colon cancer cells from radiation-induced mitotic catastrophe. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1235–G1244. doi: 10.1152/ajpgi.00037.2008. [DOI] [PubMed] [Google Scholar]
- 90.Subramaniam D, Natarajan G, Ramalingam S, Ramachandran I, May R, Queimado L, Houchen CW, Anant S. Translation inhibition during cell cycle arrest and apoptosis: Mcl-1 is a novel target for RNA binding protein CUGBP2. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1025–G1032. doi: 10.1152/ajpgi.00602.2007. [DOI] [PubMed] [Google Scholar]
- 91.Kudinov AE, Karanicolas J, Golemis EA, Boumber Y. Musashi RNA-Binding Proteins as Cancer Drivers and Novel Therapeutic Targets. Clin Cancer Res. 2017;23:2143–2153. doi: 10.1158/1078-0432.CCR-16-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nahas GR, Murthy RG, Patel SA, Ganta T, Greco SJ, Rameshwar P. The RNA-binding protein Musashi 1 stabilizes the oncotachykinin 1 mRNA in breast cancer cells to promote cell growth. FASEB J. 2016;30:149–159. doi: 10.1096/fj.15-278770. [DOI] [PubMed] [Google Scholar]
- 93.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 94.Wolfe AR, Ernlund A, McGuinness W, Lehmann C, Carl K, Balmaceda N, Neufeld KL. Suppression of intestinal tumorigenesis in Apc mutant mice upon Musashi-1 deletion. J Cell Sci. 2017;130:805–813. doi: 10.1242/jcs.197574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Baou M, Norton JD, Murphy JJ. AU-rich RNA binding proteins in hematopoiesis and leukemogenesis. Blood. 2011;118:5732–5740. doi: 10.1182/blood-2011-07-347237. [DOI] [PubMed] [Google Scholar]
- 96.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bikkavilli RK, Malbon CC. Dishevelled-KSRP complex regulates Wnt signaling through post-transcriptional stabilization of beta-catenin mRNA. J Cell Sci. 2010;123:1352–1362. doi: 10.1242/jcs.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.David Gerecht PS, Taylor MA, Port JD. Intracellular localization and interaction of mRNA binding proteins as detected by FRET. BMC Cell Biol. 2010;11:69. doi: 10.1186/1471-2121-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.von Roretz C, Di Marco S, Mazroui R, Gallouzi IE. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip Rev RNA. 2011;2:336–347. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- 101.Choi S, Sa M, Cho N, Kim KK, Park SH. Rbfox2 dissociation from stress granules suppresses cancer progression. Exp Mol Med. 2019;51:1–12. doi: 10.1038/s12276-019-0246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaehler C, Isensee J, Hucho T, Lehrach H, Krobitsch S. 5-Fluorouracil affects assembly of stress granules based on RNA incorporation. Nucleic Acids Res. 2014;42:6436–6447. doi: 10.1093/nar/gku264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li XY, Hu Y, Li NS, Wan JH, Zhu Y, Lu NH. RACK1 Acts as a Potential Tumor Promoter in Colorectal Cancer. Gastroenterol Res Pract. 2019;2019:5625026. doi: 10.1155/2019/5625026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Long J, Yin Y, Guo H, Li S, Sun Y, Zeng C, Zhu W. The mechanisms and clinical significance of PDCD4 in colorectal cancer. Gene. 2019;680:59–64. doi: 10.1016/j.gene.2018.09.034. [DOI] [PubMed] [Google Scholar]
- 105.Ishihama K, Yamakawa M, Semba S, Takeda H, Kawata S, Kimura S, Kimura W. Expression of HDAC1 and CBP/p300 in human colorectal carcinomas. J Clin Pathol. 2007;60:1205–1210. doi: 10.1136/jcp.2005.029165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lazarova DL, Chiaro C, Wong T, Drago E, Rainey A, O'Malley S, Bordonaro M. CBP Activity Mediates Effects of the Histone Deacetylase Inhibitor Butyrate on WNT Activity and Apoptosis in Colon Cancer Cells. J Cancer. 2013;4:481–490. doi: 10.7150/jca.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 108.Lee DH, Won HR, Ryu HW, Han JM, Kwon SH. The HDAC6 inhibitor ACY-1215 enhances the anticancer activity of oxaliplatin in colorectal cancer cells. Int J Oncol. 2018;53:844–854. doi: 10.3892/ijo.2018.4405. [DOI] [PubMed] [Google Scholar]
- 109.Jedrusik-Bode M, Studencka M, Smolka C, Baumann T, Schmidt H, Kampf J, Paap F, Martin S, Tazi J, Müller KM, Krüger M, Braun T, Bober E. The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J Cell Sci. 2013;126:5166–5177. doi: 10.1242/jcs.130708. [DOI] [PubMed] [Google Scholar]
- 110.Qi J, Cui C, Deng Q, Wang L, Chen R, Zhai D, Xie L, Yu J. Downregulated SIRT6 and upregulated NMNAT2 are associated with the presence, depth and stage of colorectal cancer. Oncol Lett. 2018;16:5829–5837. doi: 10.3892/ol.2018.9400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tian J, Yuan L. Sirtuin 6 inhibits colon cancer progression by modulating PTEN/AKT signaling. Biomed Pharmacother. 2018;106:109–116. doi: 10.1016/j.biopha.2018.06.070. [DOI] [PubMed] [Google Scholar]
- 112.Geng CH, Zhang CL, Zhang JY, Gao P, He M, Li YL. Overexpression of Sirt6 is a novel biomarker of malignant human colon carcinoma. J Cell Biochem. 2018;119:3957–3967. doi: 10.1002/jcb.26539. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Y, Nie L, Xu K, Fu Y, Zhong J, Gu K, Zhang L. SIRT6, a novel direct transcriptional target of FoxO3a, mediates colon cancer therapy. Theranostics. 2019;9:2380–2394. doi: 10.7150/thno.29724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yefi R, Ponce DP, Niechi I, Silva E, Cabello P, Rodriguez DA, Marcelain K, Armisen R, Quest AF, Tapia JC. Protein kinase CK2 promotes cancer cell viability via up-regulation of cyclooxygenase-2 expression and enhanced prostaglandin E2 production. J Cell Biochem. 2011;112:3167–3175. doi: 10.1002/jcb.23247. [DOI] [PubMed] [Google Scholar]
- 115.Lee WH, Lee HH, Vo MT, Kim HJ, Ko MS, Im YC, Min YJ, Lee BJ, Cho WJ, Park JW. Casein kinase 2 regulates the mRNA-destabilizing activity of tristetraprolin. J Biol Chem. 2011;286:21577–21587. doi: 10.1074/jbc.M110.201137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Koch S, Capaldo CT, Hilgarth RS, Fournier B, Parkos CA, Nusrat A. Protein kinase CK2 is a critical regulator of epithelial homeostasis in chronic intestinal inflammation. Mucosal Immunol. 2013;6:136–145. doi: 10.1038/mi.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tapia JC, Torres VA, Rodriguez DA, Leyton L, Quest AF. Casein kinase 2 (CK2) increases survivin expression via enhanced beta-catenin-T cell factor/lymphoid enhancer binding factor-dependent transcription. Proc Natl Acad Sci USA. 2006;103:15079–15084. doi: 10.1073/pnas.0606845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim HJ, Han YS, Lee JH, Lee SH. Casein Kinase 2α Enhances 5-Fluorouracil Resistance in Colorectal Cancer Cells by Inhibiting Endoplasmic Reticulum Stress. Anticancer Res. 2020;40:1419–1426. doi: 10.21873/anticanres.14083. [DOI] [PubMed] [Google Scholar]
- 119.Mathioudaki K, Papadokostopoulou A, Scorilas A, Xynopoulos D, Agnanti N, Talieri M. The PRMT1 gene expression pattern in colon cancer. Br J Cancer. 2008;99:2094–2099. doi: 10.1038/sj.bjc.6604807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsai WC, Reineke LC, Jain A, Jung SY, Lloyd RE. Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1. J Biol Chem. 2017;292:18886–18896. doi: 10.1074/jbc.M117.800706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang F, He L, Huangyang P, Liang J, Si W, Yan R, Han X, Liu S, Gui B, Li W, Miao D, Jing C, Liu Z, Pei F, Sun L, Shang Y. JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 2014;12:e1001819. doi: 10.1371/journal.pbio.1001819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Francipane MG, Lagasse E. mTOR pathway in colorectal cancer: an update. Oncotarget. 2014;5:49–66. doi: 10.18632/oncotarget.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Heberle AM, Prentzell MT, van Eunen K, Bakker BM, Grellscheid SN, Thedieck K. Molecular mechanisms of mTOR regulation by stress. Mol Cell Oncol. 2015;2:e970489. doi: 10.4161/23723548.2014.970489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Papadatos-Pastos D, Rabbie R, Ross P, Sarker D. The role of the PI3K pathway in colorectal cancer. Crit Rev Oncol Hematol. 2015;94:18–30. doi: 10.1016/j.critrevonc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 125.Pranteda A, Piastra V, Stramucci L, Fratantonio D, Bossi G. The p38 MAPK Signaling Activation in Colorectal Cancer upon Therapeutic Treatments. Int J Mol Sci. 2020;21:2773. doi: 10.3390/ijms21082773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang YN, Lu YX, Liu J, Jin Y, Bi HC, Zhao Q, Liu ZX, Li YQ, Hu JJ, Sheng H, Jiang YM, Zhang C, Tian F, Chen Y, Pan ZZ, Chen G, Zeng ZL, Liu KY, Ogasawara M, Yun JP, Ju HQ, Feng JX, Xie D, Gao S, Jia WH, Kopetz S, Xu RH, Wang F. AMPKα1 confers survival advantage of colorectal cancer cells under metabolic stress by promoting redox balance through the regulation of glutathione reductase phosphorylation. Oncogene. 2020;39:637–650. doi: 10.1038/s41388-019-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guo B, Han X, Tkach D, Huang SG, Zhang D. AMPK promotes the survival of colorectal cancer stem cells. Animal Model Exp Med. 2018;1:134–142. doi: 10.1002/ame2.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 129.Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, Seguin SJ, Morelli FF, Vinet J, Leo G, Pansarasa O, Cereda C, Poletti A, Alberti S, Carra S. A Surveillance Function of the HSPB8-BAG3-HSP70 Chaperone Complex Ensures Stress Granule Integrity and Dynamism. Mol Cell. 2016;63:796–810. doi: 10.1016/j.molcel.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 130.Matus S, Bosco DA, Hetz C. Autophagy meets fused in sarcoma-positive stress granules. Neurobiol Aging. 2014;35:2832–2835. doi: 10.1016/j.neurobiolaging.2014.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Minoia M, Boncoraglio A, Vinet J, Morelli FF, Brunsting JF, Poletti A, Krom S, Reits E, Kampinga HH, Carra S. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: implications for a proteasome-to-autophagy switch. Autophagy. 2014;10:1603–1621. doi: 10.4161/auto.29409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhong MA, Zhang H, Qi XY, Lu AG, You TG, Gao W, Guo XL, Zhou ZQ, Yang Y, Wang CJ. ShRNA-mediated gene silencing of heat shock protein 70 inhibits human colon cancer growth. Mol Med Rep. 2011;4:805–810. doi: 10.3892/mmr.2011.528. [DOI] [PubMed] [Google Scholar]
- 133.Soleimani A, Zahiri E, Ehtiati S, Norouzi M, Rahmani F, Fiuji H, Avan A, Ferns GA, Khazaei M, Hashemy SI, Hassanian SM. Therapeutic potency of heat-shock protein-70 in the pathogenesis of colorectal cancer: current status and perspectives. Biochem Cell Biol. 2019;97:85–90. doi: 10.1139/bcb-2018-0177. [DOI] [PubMed] [Google Scholar]
- 134.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Burada F, Nicoli ER, Ciurea ME, Uscatu DC, Ioana M, Gheonea DI. Autophagy in colorectal cancer: An important switch from physiology to pathology. World J Gastrointest Oncol. 2015;7:271–284. doi: 10.4251/wjgo.v7.i11.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Meyer H, Weihl CC. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang B, Maxwell BA, Joo JH, Gwon Y, Messing J, Mishra A, Shaw TI, Ward AL, Quan H, Sakurada SM, Pruett-Miller SM, Bertorini T, Vogel P, Kim HJ, Peng J, Taylor JP, Kundu M. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol Cell. 2019;74:742–757.e8. doi: 10.1016/j.molcel.2019.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 141.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zlobec I, Karamitopoulou E, Terracciano L, Piscuoglio S, Iezzi G, Muraro MG, Spagnoli G, Baker K, Tzankov A, Lugli A. TIA-1 cytotoxic granule-associated RNA binding protein improves the prognostic performance of CD8 in mismatch repair-proficient colorectal cancer. PLoS One. 2010;5:e14282. doi: 10.1371/journal.pone.0014282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gao X, Jiang L, Gong Y, Chen X, Ying M, Zhu H, He Q, Yang B, Cao J. Stress granule: A promising target for cancer treatment. Br J Pharmacol. 2019;176:4421–4433. doi: 10.1111/bph.14790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu Y, Pang L, Wang H, Xu C, Shah H, Guo P, Shu D, Qian SY. Specific delivery of delta-5-desaturase siRNA via RNA nanoparticles supplemented with dihomo-γ-linolenic acid for colon cancer suppression. Redox Biol. 2019;21:101085. doi: 10.1016/j.redox.2018.101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Oi N, Yuan J, Malakhova M, Luo K, Li Y, Ryu J, Zhang L, Bode AM, Xu Z, Li Y, Lou Z, Dong Z. Resveratrol induces apoptosis by directly targeting Ras-GTPase-activating protein SH3 domain-binding protein 1. Oncogene. 2015;34:2660–2671. doi: 10.1038/onc.2014.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shim JH, Su ZY, Chae JI, Kim DJ, Zhu F, Ma WY, Bode AM, Yang CS, Dong Z. Epigallocatechin gallate suppresses lung cancer cell growth through Ras-GTPase-activating protein SH3 domain-binding protein 1. Cancer Prev Res (Phila) 2010;3:670–679. doi: 10.1158/1940-6207.CAPR-09-0185. [DOI] [PubMed] [Google Scholar]
- 147.Porru M, Zizza P, Franceschin M, Leonetti C, Biroccio A. EMICORON: A multi-targeting G4 ligand with a promising preclinical profile. Biochim Biophys Acta Gen Subj. 2017;1861:1362–1370. doi: 10.1016/j.bbagen.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 148.Gho YS, Yoon WH, Chae CB. Antiplasmin activity of a peptide that binds to the receptor-binding site of angiogenin. J Biol Chem. 2002;277:9690–9694. doi: 10.1074/jbc.M105526200. [DOI] [PubMed] [Google Scholar]
- 149.Olson KA, Fett JW, French TC, Key ME, Vallee BL. Angiogenin antagonists prevent tumor growth in vivo. Proc Natl Acad Sci USA. 1995;92:442–446. doi: 10.1073/pnas.92.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sfakianos AP, Mellor LE, Pang YF, Kritsiligkou P, Needs H, Abou-Hamdan H, Désaubry L, Poulin GB, Ashe MP, Whitmarsh AJ. The mTOR-S6 kinase pathway promotes stress granule assembly. Cell Death Differ. 2018;25:1766–1780. doi: 10.1038/s41418-018-0076-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hofmann S, Cherkasova V, Bankhead P, Bukau B, Stoecklin G. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell. 2012;23:3786–3800. doi: 10.1091/mbc.E12-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang WL, Perillo W, Liou D, Marambaud P, Wang P. AMPK inhibitor compound C suppresses cell proliferation by induction of apoptosis and autophagy in human colorectal cancer cells. J Surg Oncol. 2012;106:680–688. doi: 10.1002/jso.23184. [DOI] [PubMed] [Google Scholar]
- 153.Ivanov PA, Chudinova EM, Nadezhdina ES. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp Cell Res. 2003;290:227–233. doi: 10.1016/s0014-4827(03)00290-8. [DOI] [PubMed] [Google Scholar]
- 154.Rtibi K, Grami D, Selmi S, Amri M, Sebai H, Marzouki L. Vinblastine, an anticancer drug, causes constipation and oxidative stress as well as others disruptions in intestinal tract in rat. Toxicol Rep. 2017;4:221–225. doi: 10.1016/j.toxrep.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cortes J, O'Shaughnessy J, Loesch D, Blum JL, Vahdat LT, Petrakova K, Chollet P, Manikas A, Diéras V, Delozier T, Vladimirov V, Cardoso F, Koh H, Bougnoux P, Dutcus CE, Seegobin S, Mir D, Meneses N, Wanders J, Twelves C EMBRACE (Eisai Metastatic Breast Cancer Study Assessing Physician's Choice Versus E7389) investigators. Eribulin monotherapy versus treatment of physician's choice in patients with metastatic breast cancer (EMBRACE): a phase 3 open-label randomised study. Lancet. 2011;377:914–923. doi: 10.1016/S0140-6736(11)60070-6. [DOI] [PubMed] [Google Scholar]
- 156.Perez-Garcia JM, Cortes J. The safety of eribulin for the treatment of metastatic breast cancer. Expert Opin Drug Saf. 2019;18:347–355. doi: 10.1080/14740338.2019.1608946. [DOI] [PubMed] [Google Scholar]
- 157.Kaliszczak M, van Hechanova E, Li Y, Alsadah H, Parzych K, Auner HW, Aboagye EO. The HDAC6 inhibitor C1A modulates autophagy substrates in diverse cancer cells and induces cell death. Br J Cancer. 2018;119:1278–1287. doi: 10.1038/s41416-018-0232-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Won HR, Ryu HW, Shin DH, Yeon SK, Lee DH, Kwon SH. A452, an HDAC6-selective inhibitor, synergistically enhances the anticancer activity of chemotherapeutic agents in colorectal cancer cells. Mol Carcinog. 2018;57:1383–1395. doi: 10.1002/mc.22852. [DOI] [PubMed] [Google Scholar]
- 159.Chen MC, Lin YC, Liao YH, Liou JP, Chen CH. MPT0G612, a Novel HDAC6 Inhibitor, Induces Apoptosis and Suppresses IFN-γ-Induced Programmed Death-Ligand 1 in Human Colorectal Carcinoma Cells. Cancers (Basel) 2019;11:1617. doi: 10.3390/cancers11101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jiang H, Cheng ST, Ren JH, Ren F, Yu HB, Wang Q, Huang AL, Chen J. SIRT6 Inhibitor, OSS_128167 Restricts Hepatitis B Virus Transcription and Replication Through Targeting Transcription Factor Peroxisome Proliferator-Activated Receptors α. Front Pharmacol. 2019;10:1270. doi: 10.3389/fphar.2019.01270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Damonte P, Sociali G, Parenti MD, Soncini D, Bauer I, Boero S, Grozio A, Holtey MV, Piacente F, Becherini P, Sanguineti R, Salis A, Damonte G, Cea M, Murone M, Poggi A, Nencioni A, Del Rio A, Bruzzone S. SIRT6 inhibitors with salicylate-like structure show immunosuppressive and chemosensitizing effects. Bioorg Med Chem. 2017;25:5849–5858. doi: 10.1016/j.bmc.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 162.Khan RI, Nirzhor SSR, Akter R. A Review of the Recent Advances Made with SIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets. Biomolecules. 2018;8:44. doi: 10.3390/biom8030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Krisenko MO, Higgins RL, Ghosh S, Zhou Q, Trybula JS, Wang WH, Geahlen RL. Syk Is Recruited to Stress Granules and Promotes Their Clearance through Autophagy. J Biol Chem. 2015;290:27803–27815. doi: 10.1074/jbc.M115.642900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR) Wiley Interdiscip Rev RNA. 2020;11:e1581. doi: 10.1002/wrna.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang J, Hjelmeland AB, Nabors LB, King PH. Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells. Cancer Biol Ther. 2019;20:979–988. doi: 10.1080/15384047.2019.1591673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Blanco FF, Preet R, Aguado A, Vishwakarma V, Stevens LE, Vyas A, Padhye S, Xu L, Weir SJ, Anant S, Meisner-Kober N, Brody JR, Dixon DA. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget. 2016;7:74043–74058. doi: 10.18632/oncotarget.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Durko L, Malecka-Panas E. Lifestyle Modifications and Colorectal Cancer. Curr Colorectal Cancer Rep. 2014;10:45–54. doi: 10.1007/s11888-013-0203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Valentin-Vega YA, Wang YD, Parker M, Patmore DM, Kanagaraj A, Moore J, Rusch M, Finkelstein D, Ellison DW, Gilbertson RJ, Zhang J, Kim HJ, Taylor JP. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci Rep. 2016;6:25996. doi: 10.1038/srep25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111:1051–1061. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- 170.Sama RR, Ward CL, Kaushansky LJ, Lemay N, Ishigaki S, Urano F, Bosco DA. FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J Cell Physiol. 2013;228:2222–2231. doi: 10.1002/jcp.24395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hamdollah Zadeh MA, Amin EM, Hoareau-Aveilla C, Domingo E, Symonds KE, Ye X, Heesom KJ, Salmon A, D'Silva O, Betteridge KB, Williams AC, Kerr DJ, Salmon AH, Oltean S, Midgley RS, Ladomery MR, Harper SJ, Varey AH, Bates DO. Alternative splicing of TIA-1 in human colon cancer regulates VEGF isoform expression, angiogenesis, tumour growth and bevacizumab resistance. Mol Oncol. 2015;9:167–178. doi: 10.1016/j.molonc.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang Q, Zhu J, Wang YW, Dai Y, Wang YL, Wang C, Liu J, Baker A, Colburn NH, Yang HS. Tumor suppressor Pdcd4 attenuates Sin1 translation to inhibit invasion in colon carcinoma. Oncogene. 2017;36:6225–6234. doi: 10.1038/onc.2017.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ramalingam S, Ramamoorthy P, Subramaniam D, Anant S. Reduced Expression of RNA Binding Protein CELF2, a Putative Tumor Suppressor Gene in Colon Cancer. Immunogastroenterology. 2012;1:27–33. doi: 10.7178/ig.1.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li N, Chen M, Cao Y, Li H, Zhao J, Zhai Z, Ren F, Li K. Bcl-2-associated athanogene 3(BAG3) is associated with tumor cell proliferation, migration, invasion and chemoresistance in colorectal cancer. BMC Cancer. 2018;18:793. doi: 10.1186/s12885-018-4657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jeon JY, Lee JS, Park ER, Shen YN, Kim MY, Shin HJ, Joo HY, Cho EH, Moon SM, Shin US, Park SH, Han CJ, Choi DW, Gu MB, Kim SB, Lee KH. Protein arginine methyltransferase 5 is implicated in the aggressiveness of human hepatocellular carcinoma and controls the invasive activity of cancer cells. Oncol Rep. 2018;40:536–544. doi: 10.3892/or.2018.6402. [DOI] [PubMed] [Google Scholar]
- 176.Wang Q, Tan R, Zhu X, Zhang Y, Tan Z, Su B, Li Y. Oncogenic K-ras confers SAHA resistance by up-regulating HDAC6 and c-myc expression. Oncotarget. 2016;7:10064–10072. doi: 10.18632/oncotarget.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lin KY, Tai C, Hsu JC, Li CF, Fang CL, Lai HC, Hseu YC, Lin YF, Uen YH. Overexpression of nuclear protein kinase CK2 α catalytic subunit (CK2α) as a poor prognosticator in human colorectal cancer. PLoS One. 2011;6:e17193. doi: 10.1371/journal.pone.0017193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zou Y, Chen Z, He X, He X, Wu X, Chen Y, Wu X, Wang J, Lan P. High expression levels of unc-51-like kinase 1 as a predictor of poor prognosis in colorectal cancer. Oncol Lett. 2015;10:1583–1588. doi: 10.3892/ol.2015.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Choi EJ, Lee JH, Kim MS, Song SY, Yoo NJ, Lee SH. Intratumoral Heterogeneity of Somatic Mutations for NRIP1, DOK1, ULK1, ULK2, DLGAP3, PARD3 and PRKCI in Colon Cancers. Pathol Oncol Res. 2018;24:827–832. doi: 10.1007/s12253-017-0297-0. [DOI] [PubMed] [Google Scholar]
- 180.Yamamoto S, Tomita Y, Hoshida Y, Sakon M, Kameyama M, Imaoka S, Sekimoto M, Nakamori S, Monden M, Aozasa K. Expression of valosin-containing protein in colorectal carcinomas as a predictor for disease recurrence and prognosis. Clin Cancer Res. 2004;10:651–657. doi: 10.1158/1078-0432.ccr-1576-03. [DOI] [PubMed] [Google Scholar]
- 181.Jin H, Gong W, Zhang C, Wang S. Epigallocatechin gallate inhibits the proliferation of colorectal cancer cells by regulating Notch signaling. Onco Targets Ther. 2013;6:145–153. doi: 10.2147/OTT.S40914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Heerma van Voss MR, Vesuna F, Trumpi K, Brilliant J, Berlinicke C, de Leng W, Kranenburg O, Offerhaus GJ, Bürger H, van der Wall E, van Diest PJ, Raman V. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget. 2015;6:28312–28326. doi: 10.18632/oncotarget.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang XW, Zhang YJ. Targeting mTOR network in colorectal cancer therapy. World J Gastroenterol. 2014;20:4178–4188. doi: 10.3748/wjg.v20.i15.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ng K, Tabernero J, Hwang J, Bajetta E, Sharma S, Del Prete SA, Arrowsmith ER, Ryan DP, Sedova M, Jin J, Malek K, Fuchs CS. Phase II study of everolimus in patients with metastatic colorectal adenocarcinoma previously treated with bevacizumab-, fluoropyrimidine-, oxaliplatin-, and irinotecan-based regimens. Clin Cancer Res. 2013;19:3987–3995. doi: 10.1158/1078-0432.CCR-13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Spindler KL, Sorensen MM, Pallisgaard N, Andersen RF, Havelund BM, Ploen J, Lassen U, Jakobsen AK. Phase II trial of temsirolimus alone and in combination with irinotecan for KRAS mutant metastatic colorectal cancer: outcome and results of KRAS mutational analysis in plasma. Acta Oncol. 2013;52:963–970. doi: 10.3109/0284186X.2013.776175. [DOI] [PubMed] [Google Scholar]
- 186.Wang L, Hu T, Shen J, Zhang L, Chan RL, Lu L, Li M, Cho CH, Wu WK. Dihydrotanshinone I induced apoptosis and autophagy through caspase dependent pathway in colon cancer. Phytomedicine. 2015;22:1079–1087. doi: 10.1016/j.phymed.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 187.Einzig AI, Neuberg D, Wiernik PH, Grochow LB, Ramirez G, O'Dwyer PJ, Petrelli NJ. Phase II Trial of Paclitaxel in Patients with Advanced Colon Cancer Previously Untreated with Cytotoxic Chemotherapy: An Eastern Cooperative Oncology Group Trial (PA286) Am J Ther. 1996;3:750–754. doi: 10.1097/00045391-199611000-00003. [DOI] [PubMed] [Google Scholar]
- 188.Auyeung KK, Law PC, Ko JK. Combined therapeutic effects of vinblastine and Astragalus saponins in human colon cancer cells and tumor xenograft via inhibition of tumor growth and proangiogenic factors. Nutr Cancer. 2014;66:662–674. doi: 10.1080/01635581.2014.894093. [DOI] [PubMed] [Google Scholar]