Figure 4.
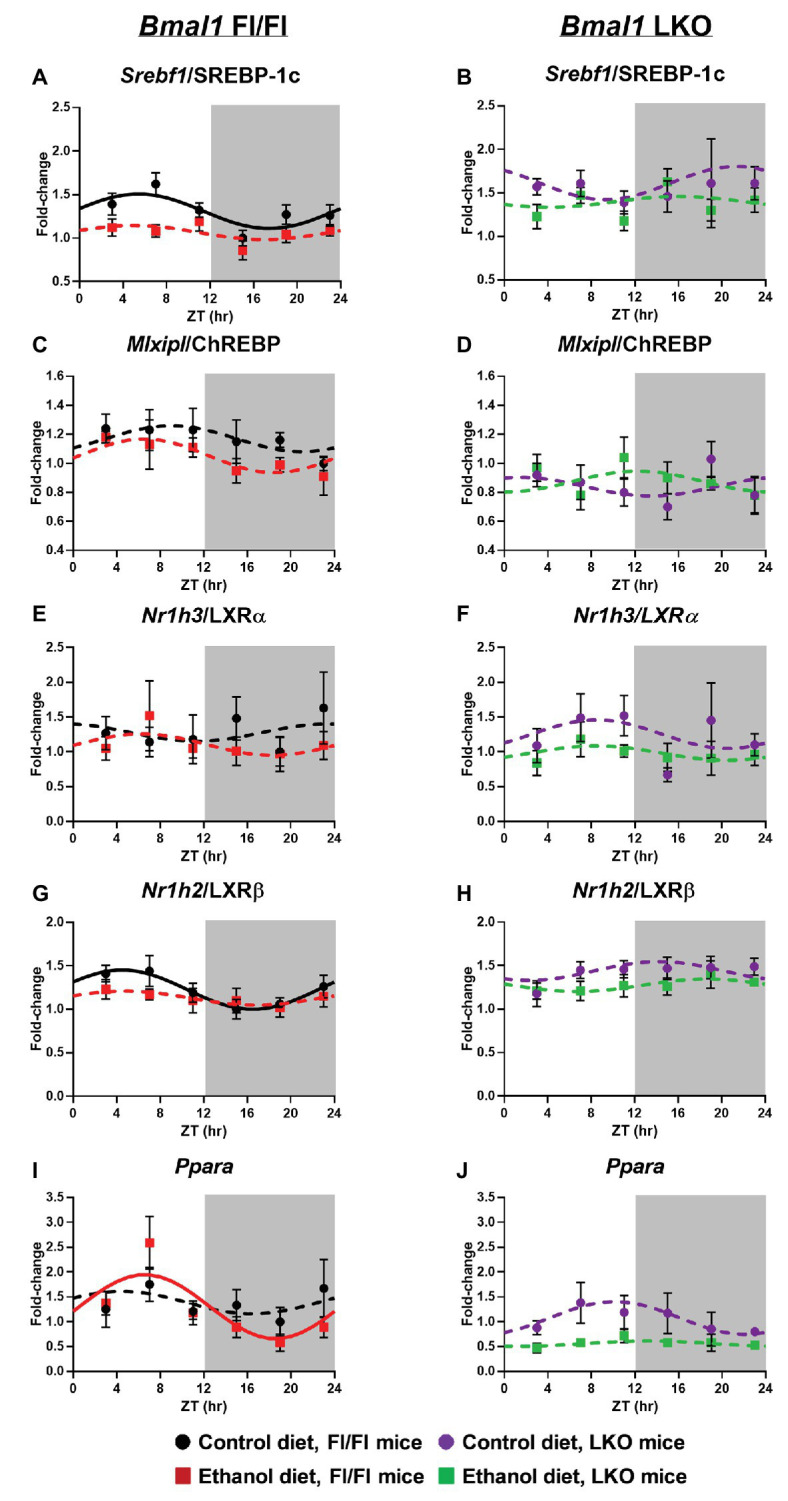
Chronic alcohol and liver clock disruption alter diurnal mRNA rhythms of lipid metabolism transcription factors in the liver. Diurnal mRNA profiles of Sterol regulatory element binding transcription factor 1 (Srebf1/SREBP-1C; A,B), MLX-interacting protein-like/Carbohydrate-responsive element-binding protein (Mlxipl/ChREBP; C,D), Nuclear receptor subfamily 1, group H, member 3/Liver X receptor alpha (Nr1h3/ LXRα; E,F), Nuclear receptor subfamily 1, group H, member 2/Liver X receptor beta (Nr1h2/LXRβ; G,H), Peroxisome proliferator-activated receptor alpha (Pparα; I,J) were measured in livers of control-fed (black) and alcohol-fed (red) Bmal1 Flox/Flox (Fl/Fl) and control-fed (purple) and alcohol-fed (green) Bmal1 liver-specific knockout (LKO) mice at ZT 3, 7, 11, 15, 19, and 23 (ZT 0: lights on/inactive period, ZT 12: lights off/active period, gray area) by RT-PCR. Data are presented as a fold-change to the Bmal1 Fl/Fl control diet trough time-point and normalized to Ppia. Cosinor analysis was performed using a nonlinear regression module in SPSS. Data are expressed as mean ± SEM for n = 4–8 mice/genotype/diet/time point. Solid lines indicate rhythmic mRNA levels and a significant cosine fit, whereas dashed lines indicate arrhythmicity and a non-significant cosine fit. Results for Cosinor and ANOVA analyses are provided in Supplementary Tables 3 and 4, respectively.
