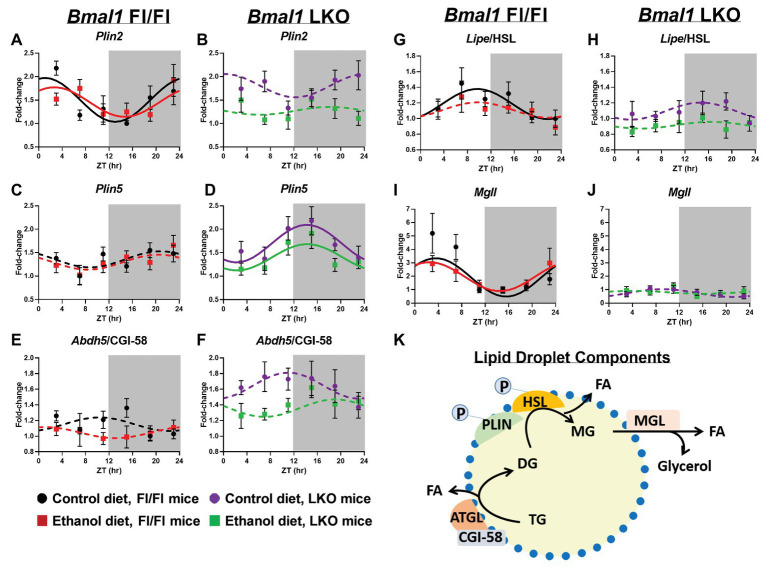
Figure 8.
Chronic alcohol and liver clock disruption alter diurnal mRNA rhythms of lipid droplet-associated genes. Diurnal mRNA profiles of lipid droplet genes Perilipin 2 (Plin2; A,B), Perilipin 5 (Plin5; C,D), Abhydrolase domain containing 5/Comparative Gene Interaction-58 (Abdh5/CGI-58; E,F), Hormone sensitive lipase (Lipe/HSL; G,H), and Monoacylglycerol lipase (Mgll; I,J) were measured in livers of control-fed (black) and alcohol-fed (red) Bmal1 Flox/Flox (Fl/Fl) and control-fed (purple) and alcohol-fed (green) Bmal1 liver-specific knockout (LKO) mice at ZT 3, 7, 11, 15, 19, and 23 (ZT 0: lights on/inactive period, ZT12: lights off/active period, gray area) by RT-PCR. Data are presented as a fold-change to the Bmal1 Fl/Fl control diet trough time-point and normalized to Ppia. Data are expressed as mean ± SEM for n = 4–8 mice/genotype/diet/time point. Solid lines indicate rhythmic mRNA levels and a significant cosine fit, whereas dashed lines indicate arrhythmicity and a non-significant cosine fit. Diagram of a lipid droplet and lipid droplet-associated turnover proteins and enzymes (K). After phosphorylation, PLIN recruits lipases to the surface of the lipid droplet to initiate TG breakdown to glycerol and FA. Results for Cosinor and ANOVA analyses are provided in Supplementary Tables 4 and 7, respectively.