Itaconate is a dicarboxylic acid that inhibits the isocitrate lyase enzyme of the bacterial glyoxylate shunt. Activated macrophages have been shown to produce itaconate, suggesting that these immune cells may employ this metabolite as a weapon against invading bacteria. Here, we demonstrate that in vitro, itaconate can exhibit bactericidal effects under acidic conditions similar to the pH of a macrophage phagosome. In parallel, successful pathogens, including Salmonella, have acquired a genetic operon encoding itaconate degradation proteins, which are induced heavily in macrophages.
KEYWORDS: Salmonella, acid resistance, biosensors, gene regulation, host-pathogen interactions, itaconate, macrophages, metabolism, pathogenicity islands, promoters
ABSTRACT
Itaconate is a dicarboxylic acid that inhibits the isocitrate lyase enzyme of the bacterial glyoxylate shunt. Activated macrophages have been shown to produce itaconate, suggesting that these immune cells may employ this metabolite as a weapon against invading bacteria. Here, we demonstrate that in vitro, itaconate can exhibit bactericidal effects under acidic conditions similar to the pH of a macrophage phagosome. In parallel, successful pathogens, including Salmonella, have acquired a genetic operon encoding itaconate degradation proteins, which are induced heavily in macrophages. We characterized the regulation of this operon by the neighboring gene ripR in specific response to itaconate. Moreover, we developed an itaconate biosensor based on the operon promoter that can detect itaconate in a semiquantitative manner and, when combined with the ripR gene, is sufficient for itaconate-regulated expression in Escherichia coli. Using this biosensor with fluorescence microscopy, we observed bacteria responding to itaconate in the phagosomes of macrophages and provide additional evidence that gamma interferon stimulates macrophage itaconate synthesis and that J774 mouse macrophages produce substantially more itaconate than the human THP-1 monocyte cell line. In summary, we examined the role of itaconate as an antibacterial metabolite in mouse and human macrophages, characterized the regulation of Salmonella’s defense against it, and developed it as a convenient itaconate biosensor and inducible promoter system.
INTRODUCTION
The mammalian immune system includes a multitude of weapons to defend against invading microbes, and successful pathogens have evolved a plethora of mechanisms to evade, manipulate, or even benefit from these immune responses. One such pathogen, Salmonella enterica serovar Typhimurium (here referred to as Salmonella), has acquired a number of Salmonella pathogenicity islands (SPI) that support its survival inside a host organism. For instance, Salmonella employs SPI-1 to invade nonphagocytic cells, and SPI-2 allows the bacteria to survive intracellularly, including in macrophages, which is important for Salmonella virulence (1–4). These traits allow Salmonella to invade the gut epithelium and induce intestinal inflammation resulting in the characteristic gastroenteritis disease. Moreover, the induced inflammation is not merely a threat that Salmonella must survive, but it has adapted to thrive in the oxidative environment of the inflamed intestine and utilize inflammation-derived metabolites to outcompete resident microbiota (5–7).
Itaconate (2-methylenesuccinic acid, 2-methylidenebutanedioic acid) is a metabolite originally recognized in fungal species such as Aspergillus terreus and produced commercially for use in polymer production (8–10). As an unsaturated dicarboxylate that is somewhat similar in structure to succinate, itaconate is a potent inhibitor of the glyoxylate shunt enzyme AceA (isocitrate lyase) (11–13). As such, itaconate inhibits bacterial growth on carbon sources such as acetate and fatty acids, conditions that necessitate the glyoxylate shunt to generate the 4-carbon skeletons that are critical for amino acid biosynthesis and central metabolism. Interestingly, it has been demonstrated that activated macrophages employ the IRG1 protein to produce itaconate, with higher concentrations produced by mouse macrophages than by human ones (13–15). Moreover, IRG1 closely associates with vesicles containing Legionella pneumophila, and itaconate showed bactericidal activity against this pathogen in vitro (15). Itaconate was also found to inhibit Salmonella growth by reducing medium pH, and itaconate levels in Salmonella-infected mice correlated with splenomegaly (16). Cumulatively, these works suggest that itaconate acts as a weaponized metabolite that the immune system employs to inhibit the growth of, or kill, invading bacteria.
If itaconate is an immune-derived antibacterial metabolite, then it follows logically that successful pathogens must have methods to evade its effects. Indeed, an operon has been identified in Yersinia (ripABC, for “required for intracellular proliferation”) that encodes three enzymes catalyzing the ATP/succinyl coenzyme A (succinyl-CoA)-dependent degradation of itaconate into pyruvate and acetyl-CoA (17). The operon is not restricted to Yersinia, and a variety of other bacteria, including Pseudomonas, harbor homologs or functional analogs of these enzymes. Several lineages of Salmonella enterica harbor a cluster of genes (e.g., genes STM3120 to STM3117 in S. Typhimurium strain LT2) within SPI-13 that we refer to here as the itaconate response operon (IRO). Interestingly, the IRO genes of Salmonella have been shown to be induced heavily in macrophages but not under any other condition tested, supporting a role in degrading macrophage-produced itaconate (18–20). High-throughput screens have suggested that genes from this operon are important for Salmonella survival in mice (21–23). Furthermore, it has also been shown that SPI-13 is present in many generalist S. enterica serovars but not in some human-restricted serovars of Salmonella (e.g., serovars Typhi and Paratyphi A, which harbor SPI-8), possibly due to the reduced itaconate synthesis by human macrophages (24).
In this work, we show that itaconate is bactericidal at low but not neutral pH and elucidate the regulation of the Salmonella IRO and its induction in mouse and human macrophages. We show that the promoter of the IRO (PIRO) is specifically induced by itaconate and that the LysR family transcriptional regulator encoded by the upstream gene, STM3121 (which we propose to name ripR), is both necessary and sufficient for this induction. Furthermore, using PIRO with a green fluorescent protein (GFP) reporter, we develop a semiquantitative itaconate biosensor and employ it to show that the IRO is induced heavily in the J774 mouse macrophage cell line but requires gamma interferon (IFN-γ) stimulation to show a detectable response in the THP-1 human monocyte cell line.
(This work was submitted to an online preprint archive [25].)
RESULTS
The Salmonella IRO is induced specifically by itaconate in a RipR-dependent manner.
The Salmonella pathogenicity island-13 includes genes encoding RipC/Ccl (STM3120 in strain LT2), RipB/Ich (STM3119), and RipA/Ict (STM3118), whose homologs have been demonstrated to degrade itaconate into pyruvate and acetyl-CoA (17). This operon, which we refer to as the itaconate response operon (IRO), appears to also include the STM3117 gene (encoding a predicted glyoxalase domain-containing protein) and is adjacent to the STM3121 (ripR) gene on the reverse DNA strand (Fig. 1).
FIG 1.
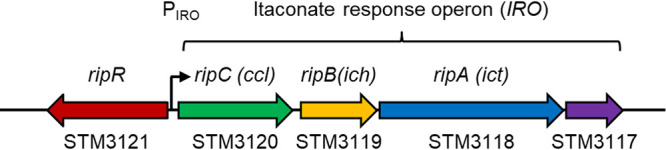
Overview of the itaconate response operon (IRO) and the itaconate-responsive regulator ripR. Salmonella gene loci based on the genome of Salmonella Typhimurium LT2 are shown at the bottom. Identities of ccl, ich, and ict are shown at the top as previously reported (17). STM3117 appears to be located in the same operon; however, its function remains unknown. ripR, STM3121; IRO, itaconate response operon (STM3120 to STM3117); PIRO, IRO promoter.
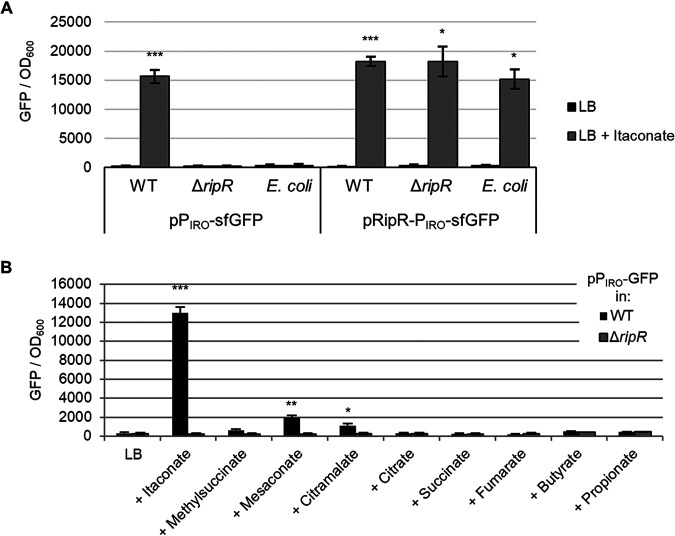
The IRO is strongly induced in cultured mouse macrophages but, according to expression data from SalCom and other published work, is seemingly independent of known virulence regulators (PhoP, RpoS, SsrA/B, OmpR, SlyA, etc.). We hypothesized that itaconate may act as an inducer of IRO expression via the adjacent LysR family regulator encoded by STM3121. To assess this, we constructed a plasmid-borne fusion of the operon’s promoter (PIRO) to superfolder GFP (sfGFP) as a reporter (26). Indeed, we found that the PIRO promoter was induced highly in the presence of itaconate and that this response was entirely dependent on the presence of RipR, as neither a Salmonella ripR deletion mutant nor the same reporter plasmid in Escherichia coli K-12 (which does not encode ripR) showed induction (Fig. 2A). In contrast, when the ripR gene was included on the reporter plasmid, itaconate-induced PIRO expression was restored in both the ΔripR Salmonella strain and in E. coli. These data demonstrate not only that PIRO is induced in response to itaconate but also that the neighboring gene ripR is both necessary and sufficient for this induction.
FIG 2.
RipR is necessary and sufficient to induce PIRO expression in response to itaconate. Expression of PIRO-sfGFP in wild-type (WT) or ripR knockout (ΔripR) Salmonella or in E. coli. Figures show GFP fluorescence normalized to optical density at 600 nm (OD600) after 16 h of growth in LB alone or supplemented with 0.2% itaconate (A) or similar metabolites (B). Data are the averages from at least three biological replicates, and error bars show one standard deviation. pPIRO, plasmid-borne transcriptional fusion of PIRO to sfGFP; pRipR-PIRO, pPIRO with the ripR gene and its native promoter included on the plasmid. A Games-Howell analysis of variance (ANOVA) was conducted comparing with and without itaconate for each strain (A) or comparing WT to ΔripR (B) for each added metabolite. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To assess if the IRO promoter is induced specifically by itaconate, we examined induction of the PIRO reporter plasmid in media supplemented with a panel of similar metabolites. While mesaconate, citramalate, and methylsuccinate (in order of induction strength) slightly induced expression, induction by itaconate was drastically more pronounced, suggesting that it is the principal inducer (Fig. 2B). Notably, similar results were obtained in complex media (LB) and in morpholinepropanesulfonic acid (MOPS) minimal medium with either glucose or glycerol as a carbon source, suggesting that the induction only requires itaconate and not additional factors in the media (see Fig. S1 in the supplemental material). Furthermore, induction by itaconate occurred in a dose-dependent manner, indicating that the reporter can be used to semiquantitatively assess itaconate concentrations encountered by the bacteria (see Fig. S2).
Itaconate import is independent of the dicarboxylate transporter DctA.
Another important question was whether itaconate required the primary aerobic dicarboxylate transporter, DctA, for import into the cell. We compared itaconate-mediated induction of the PIRO-sfGFP reporter in wild-type and ΔdctA Salmonella. Interestingly, we found that the IRO promoter was still heavily induced in the dctA mutant in the presence of itaconate (see Fig. S3). This demonstrates that the DctA dicarboxylate transporter is not required for itaconate-mediated induction of PIRO, suggesting that itaconate import is independent of dctA.
Itaconate is bactericidal at low pH.
Previous work has demonstrated that itaconate can inhibit the function of the glyoxylate shunt enzyme AceA and act as a bacteriostatic agent when bacteria rely on carbon sources such as acetate that require this pathway (11–13). It has also been suggested that itaconate can inhibit bacteria by influencing medium pH, and at least one publication has demonstrated that it can have bactericidal activity (15, 16). To clarify this later phenotype, we hypothesized that the dicarboxylic acid chemistry of itaconate would allow it to act in a bactericidal fashion at low pH by acting as a proton shuttle. In brief, the carboxyl groups of itaconate (pKa 5.5 and 3.8) protonate and lose their charge at lower pH, allowing them to traverse the bacterial membrane and release the protons in the more neutral pH of the cytoplasm, potentially exacerbating acid stress (Fig. 3A).
FIG 3.
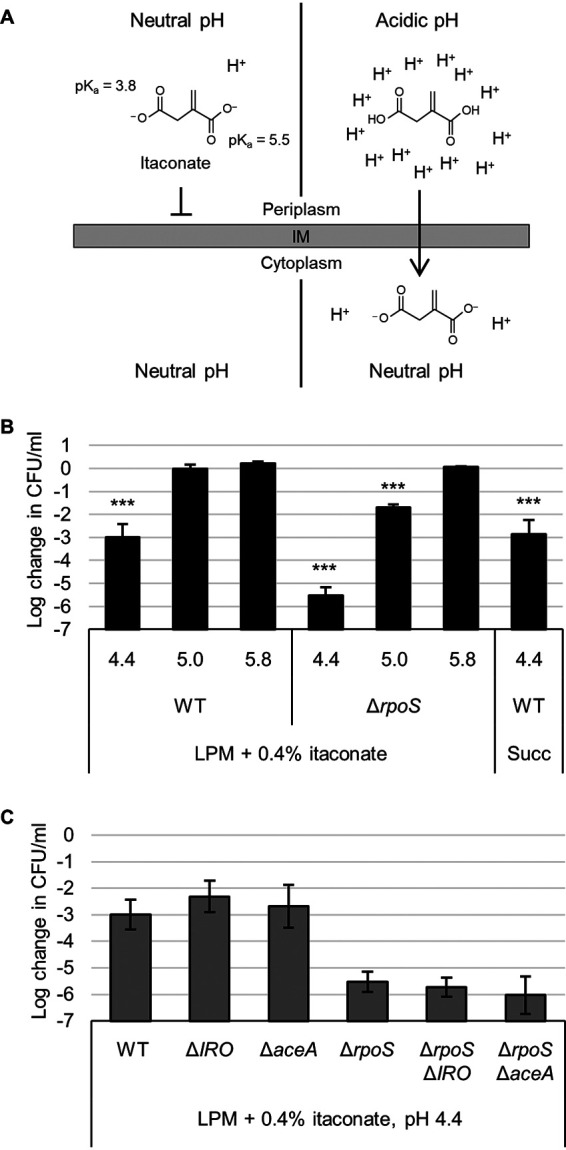
Itaconate is bactericidal at low pH. (A) At neutral pH, the charges on itaconate inhibit diffusion across a lipid membrane (left). At acidic pH, itaconate protonates to itaconic acid, which can traverse the membrane (right). In the cytoplasm, itaconic acid dissociates and releases protons, trapping it in the cell and acidifying the cytoplasm. (B) Survival (relative to 0 h time point) of wild-type (WT) or rpoS mutant Salmonella after 3 h in LPM supplemented with 0.4% itaconate or succinate (Succ) and adjusted to pH 4.4, 5.0, or 5.8 as indicated. (C) Same as in panel B but showing survival of Salmonella aceA and itaconate response operon (IRO) mutants in LPM plus itaconate medium at pH 4.4. All data are the averages from at least three biological replicates, and error bars show one standard deviation. A one-way ANOVA with Sidak’s multiple-comparison test was conducted comparing each sample to the same strain at pH 5.8 (B). WT in succinate was compared to WT at pH 5.8 in itaconate. Mutants in panel C were not significantly different than their parental strain (WT or ΔrpoS). ***, P < 0.001.
To emulate the intracellular conditions that Salmonella may encounter in a Salmonella-containing vacuole (SCV) of a macrophage, we added itaconate to low phosphate, low magnesium-containing medium (LPM) and then acidified it to pH 4.4, 5.0, or 5.8 to cover a range from the most acidic to more regular estimates of SCV pH (27–29). Indeed, we found that wild-type Salmonella showed a 1,000-fold decrease in survival after 3 h at pH 4.4 with itaconate (Fig. 3B). This lethality was alleviated at higher pH and also occurred using a similar dicarboxylic acid, succinate. Importantly, the bactericidal effect was also dependent on the presence of itaconate or succinate, as pH 4.4 LPM did not kill Salmonella in the absence of a dicarboxylic acid (see Fig. S4A). Interestingly, deletion of the entire IRO (STM3120 to STM3117) or aceA had no effect, but deletion of the general stress response sigma factor RpoS (σ32, σS) exacerbated Salmonella’s sensitivity at both pH 4.4 and 5.0 (Fig. 3C and S4B). Cumulatively, these data demonstrate that itaconate or other dicarboxylic acids can act in a bactericidal fashion under acidic conditions by exacerbating acid stress.
The Salmonella IRO does not significantly contribute to short-term survival in a mouse macrophage cell line.
The inhibitory effect of itaconate on AceA, its bactericidal activity under acidic conditions, and the synthesis of itaconate in macrophages combine to support the concept that these immune cells may be employing itaconate as an antibacterial compound. As a successful pathogen, Salmonella has adapted to survive in activated macrophages, and multiple previous works have examined how IRO genes may influence Salmonella survival and virulence (21–23). In our hands, we found no significant reduction in survival of ΔIRO or ΔripR strains in the mouse J774 macrophage cell line (see Fig. S5). When the macrophages were prestimulated with IFN-γ, there was a slight reduction in survival relative to that of the wild type, but this was not significant compared to an aceA mutant that showed no survival defect. In contrast, the growth of a phoP deletion control strain was inhibited by macrophages even without IFN-γ.
Salmonella encounters itaconate in the phagosomes of mouse and IFN-γ-stimulated human macrophages.
Multiple high-throughput studies have demonstrated that the IRO genes are induced heavily in mouse macrophages (18–20). Additional studies have identified itaconate in both mouse and human macrophages, but the mouse cells appear to produce significantly more of the metabolite (13, 14). Furthermore, it has been demonstrated that beta interferon-β (IFN-β) and IFN-γ can stimulate itaconate production in mouse macrophages (15, 30–32).
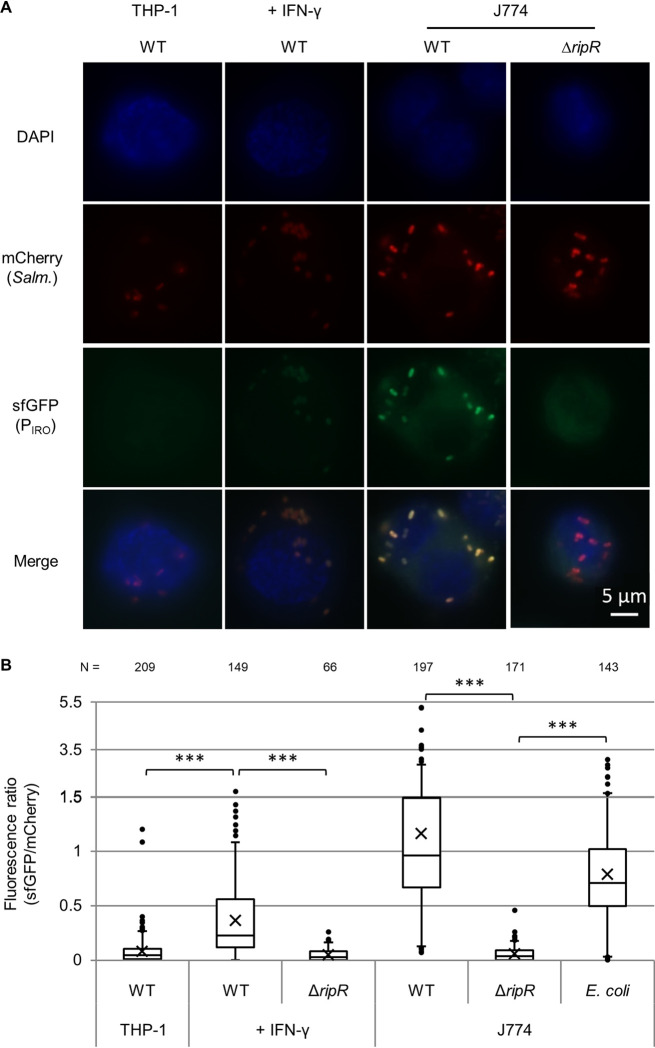
To examine itaconate levels encountered by intracellular bacteria inside macrophages, we employed our PIRO-sfGFP reporter plasmid as a biosensor. By including a constitutively expressed mCherry gene on the same plasmid, we were able to microscopically observe individual bacteria inside macrophages and obtain semiquantitative data by generating a GFP/mCherry fluorescence ratio as an indicator of PIRO induction and, accordingly, itaconate levels. Using this system, we observed strong induction of the PIRO promoter for wild-type Salmonella in J774 mouse macrophages, and this signal was absent in the ΔripR control (Fig. 4). Furthermore, the response was also observed in E. coli if RipR was encoded on the same plasmid (Fig. 4, E. coli data), demonstrating that bacteria that are poorly adapted to intracellular survival also encounter itaconate in macrophages.
FIG 4.
PIRO is activated in J774 macrophages and THP-1 macrophages stimulated with IFN-γ. Expression of PIRO-sfGFP in intracellular wild-type (WT) or ripR knockout (ΔripR) Salmonella containing the pICM-PIRO plasmid (constitutive mCherry expression). For expression in E. coli, the ripR gene was also located on the reporter plasmid. “+ IFN-γ” samples were pretreated with human IFN-γ. (A) Representative fluorescence microscopy images. (B) Relative fluorescence quantification of individual bacterial particles at 8 h postinfection. Number of bacteria quantified is indicated at the top and totaled from at least two biological replicates (≥3 for all WT Salmonella samples). Boxes indicate first and third quartiles; central line, median; X, mean; whiskers, 95th percentile; dots, all nonzero data points outside the 95th percentile. The y axis changes scale at 1.5 to better show outliers. A Games-Howell ANOVA was conducted comparing indicated samples. ***, P < 0.001.
In contrast to the mouse cell line, unstimulated human THP-1 monocytes showed negligible itaconate levels, as very few of the bacterial reporters showed any green fluorescence above background levels (Fig. 4). The bacteria did express the constitutive mCherry and were observable in the macrophages, suggesting that the lack of green fluorescence was not due to decreased bacterial survival or protein expression. Stimulation of THP-1 cells with IFN-γ (M1 activation), however, led to a significant increase in the green fluorescence of the reporter bacteria in a RipR-dependent fashion. In contrast, itaconate levels in THP-1 cells induced with interleukin 4 (IL-4) and IL-13 (M2 activation) resembled that in unstimulated cells (see Fig. S6).
DISCUSSION
In this work, we demonstrate that itaconate becomes bactericidal at acidic pH, suggesting an additional mechanism for itaconate to act as an antibacterial metabolite beyond inhibition of AceA. Thus, elevated itaconate levels in macrophages may act to inhibit bacterial metabolism while also exacerbating acid stress on microbes in the phagosome. Protonation of itaconate under acidic conditions may also grant it increased access to the bacterial cytoplasm, where deprotonation would trap the charged form close to its AceA target. This organic acid killing effect was demonstrated previously, including in a recent work showing propionate inhibition of Salmonella in mice (33, 34). Of note, we also find that bacterial killing occurs with succinate, a metabolite similar to itaconate that similarly increases in concentration in activated macrophages (35). Our findings that these dicarboxylic acids can kill Salmonella at pH 4.4 but not at higher pH may explain why Salmonella manipulates the SCV to maintain a pH closer to 5.0, in order to avoid this organic acid stress. Moreover, they imply that organic acids such as itaconate and succinate may contribute to the antibacterial activity of acidified phagosomes.
The antibacterial potential of itaconate, its synthesis in activated macrophages, and the localization of IRG1 to bacterium-containing vacuoles support its potential role as a weapon against intracellular bacteria. Here, we examined survival of Salmonella IRO or ripR deletion strains in the mouse macrophage J774 cell line but saw no significant decrease in survival, similar to a recent study examining SPI-13 in RAW264.7I macrophages (24). However, in that work, Espinoza et al. discovered that SPI-13 does play a role in Salmonella internalization into mouse—but not human—macrophages (24). Combined with previous works showing reduced survival of IRO mutants in mice, this operon may play a more significant survival role in the context of infection in animals that produce copious amounts of itaconate (20–23). Alternatively, it remains possible that the primary function of the IRO in this context is to degrade itaconate for use as a supplemental carbon source rather than to protect against its toxic potential.
We find Salmonella responds to itaconate in vitro and intracellularly by strongly inducing an operon encoding itaconate degradation proteins. This response is largely specific to itaconate and is entirely dependent on the neighboring gene (STM3121 in the prototypical Salmonella enterica serovar Typhimurium strain LT2), which we propose to rename ripR (rip operon regulator) to conform to earlier nomenclature. The ripR gene product is predicated to be a LysR family transcriptional regulator, suggesting that itaconate induces IRO expression by interacting directly with the substrate binding domain of RipR to activate it. Moreover, we find that RipR is sufficient for itaconate induction of the PIRO promoter in E. coli, demonstrating its potential as a novel inducible expression system with >50-fold higher transcription in the presence of the inexpensive and readily available inducer. A limitation of this expression system would be a requirement for growth on carbon sources independent of the glyoxylate shunt and also growth at neutral or alkaline pH, as we demonstrate that itaconate is bactericidal under acidic conditions. However, for many studies, these conditions are met, adding PIRO to the repertoire of available inducible promoter systems.
Using our PIRO-sfGFP itaconate biosensor, we showed a pronounced response in unstimulated mouse macrophages, whereas no induction was observed in the THP-1 human macrophage cell line without stimulation, suggesting that these cells are not producing itaconate to the same degree. Interferon was previously demonstrated to stimulate itaconate production in mouse macrophages, and we found that our biosensor was induced in human cells stimulated with IFN-γ (15, 30–32). Thus, while the human cell line was able to produce itaconate, it required auxiliary induction to do so and still produced less than the uninduced mouse macrophage. While it is possible that this reflects an artifact of the cell lines employed, it aligns well with previous works that quantified itaconate in both mouse and human cells (13, 14). Furthermore, Espinoza et al. recently determined that SPI-13 is abundant in generalist Salmonella serovars but not human-restricted ones (which instead harbor SPI-8), suggesting that low itaconate levels in humans render the IRO dispensable in these strains (24).
A recent study demonstrated that small molecules can inhibit the activity of the IRO proteins and sensitize Salmonella to itaconate inhibition in minimal media (36). Such drugs could also sensitize other bacteria to itaconate, including Yersinia, Pseudomonas, and Mycobacterium species, which also harbor an IRO (17, 36). Moreover, if human-restricted pathogens lack an IRO because human cells truly produce less itaconate, then they are potentially sensitive to it and itaconate itself could potentially be used as an antimicrobial against them. Our biosensor could prove invaluable in such studies for determining how much itaconate the bacteria are encountering, and the self-sufficiency of the biosensor allows it to be employed in a variety of species, providing added versatility.
In summary, here we present data that itaconate can act as a bactericidal metabolite at an acidic but physiologically relevant pH. We identify the regulatory mechanism of an itaconate response operon in Salmonella and employ its promoter as a novel biosensor of relative itaconate concentrations in macrophage phagosomes. Finally, we provide further evidence that IFN-γ stimulates itaconate synthesis in a human macrophage cell line and, moreover, that a mouse macrophage cell line already produces significant itaconate without requiring IFN-γ.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All Salmonella strains used in experiments are derivatives of Salmonella enterica subsp. enterica serovar Typhimurium (S. Typhimurium) strain 14028s. As described previously, lambda Red recombination and subsequent P22 phage transduction were used to generate all of the gene knockout mutants in this background (37–39). To allow for subsequent recombinations, the antibiotic resistance cassette was removed from the chromosome using the pCP20 plasmid encoding FLP recombinase (40). The heat-unstable pCP20 plasmid was eliminated by passaging overnight at 42°C, and loss was confirmed by antibiotic treatment.
A reporter fusion of the IRO promoter to sfGFP (PIRO-sfGFP) was generated using Gibson cloning to insert the 333 bp upstream of the STM3120 start codon (thereby including 25 bp upstream of the predicted −35 box and the 5′ untranslated region) into the pXG10sf plasmid (replacing the existing promoter) (41–43). For the reporter construct including ripR, the same region was extended to 1,570 bp upstream of the STM3120 start codon to include the entire STM3121 open reading frame (ORF) and a predicted transcriptional terminator following it. For fluorescence microscopy, constitutively expressed (PLtetO-1 promoter) mCherry was inserted into a transcriptionally independent region of the same plasmid. This variation of the plasmid was renamed “independent constitutive mCherry,” or pICM.
Metabolite induction of PIRO assay.
Induction of the Salmonella PIRO promoter was assessed using a transcriptional fusion to sfGFP in either the pXG10sf or pICM plasmid. Data from the two plasmids were combined, as the inducible region is identical and the plasmids only differ in the constitutively active mCherry expressed independently on pICM. Overnight LB cultures were used to inoculate (1/200 dilution) either LB or MOPS minimal media containing 0.2% of the indicated carbon source. Itaconate or other metabolites at neutral pH were supplemented to a concentration of 0.2%. Of note, for salts and hydrates, the final 0.2% concentration reflects the percentage of the carbon source itself; e.g., 0.2% succinate was made as 0.47% sodium succinate (dibasic) hexahydrate. Growth was conducted in a TECAN Infinite M200 plate reader at 37°C with shaking, and the optical density at 600 nm (OD600) and GFP fluorescence (475-nm and 511-nm excitation and emission wavelengths, respectively) were read every 15 min. For clarity, bar graphs show fluorescence at 16 h postinoculation. Chloramphenicol was included in all media at a concentration of 20 μg/ml to maintain the plasmids.
Acidified medium survival.
LPM was made as described previously (44, 45). Succinate or itaconate were added to 0.4%, and the pH was then adjusted to 4.4, 5.0, or 5.8, as indicated in the text and figures. LB overnight cultures were resuspended in acidified medium to an OD of 0.1 and incubated in a 37°C water bath. At designated time points, samples were taken, serially diluted, and plated for CFU enumeration.
Intramacrophage survival.
The THP-1 human monocyte cell line and the J774 mouse macrophage cell line were maintained in RPMI 1640 medium (with l-glutamine) supplemented with 10% fetal bovine serum (FBS) and 1% GlutaMAX and grown at 37°C and 5% CO2. THP-1 cells were seeded in 96-well plates at 50,000 per well with 50 nM phorbol 12-myristate 13-acetate (PMA) added to induce differentiation to adherent macrophages. After 48 h, medium was replaced with no-PMA growth medium overnight with 100 U/ml human IFN-γ or IL-4 and IL-13 added. For J774 macrophages, the cells were seeded in 96-well plates at 50,000 per well overnight with 100 U/ml mouse IFN-γ added if indicated in the text. Salmonella cells in RPMI were added onto seeded cells at a multiplicity of infection (MOI) of approximately 20:1 and centrifuged for 10 min at 1,000 rpm to maximize cell contact. After centrifuging, the samples were incubated at 37°C and 5% CO2 (time zero). After 30 min, cells were washed three times with phosphate-buffered saline (PBS) followed by fresh medium containing 100 μg/ml gentamicin to kill extracellular Salmonella. At 2 h, the medium was replaced with medium containing gentamicin at 10 μg/ml. At designated time points, intracellular bacteria were recovered using PBS containing 1% Triton X-100 and vigorous pipetting. Samples were serially diluted, and five 10-μl spots were plated for CFU counting. Each sample included three separate wells as technical replicates (a total of 15 × 10-μl spots counted per biological replicate).
Fluorescence microscopy.
Fluorescence microscopy was conducted similarly to the macrophage survival assay with some exceptions. Cells were seeded in 24-well plates containing glass coverslips at 125,000 per well. Bacteria were infected at an MOI of approximately 100 to maximize the instances of macrophages containing bacteria. At designated time points, the medium was removed and cells were washed three times with PBS. They were then fixed for 10 min at room temperature in PBS plus 4% paraformaldehyde (PFA). Following three more PBS washes, the cells were permeabilized for 10 min in PBS plus 0.2% Triton X-100 plus 1% bovine serum albumin (BSA). Coverslips were washed again, mounted on slides using 3 μl mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI), and allowed to dry overnight in the dark. Slides where viewed using a Zeiss Observer.z1 microscope using a 100× oil immersion lens objective and the Zeiss Zen microscopy software. Images were taken with a Zeiss AxioCam 506 mono camera mounted on the microscope. For all samples, a 2-s exposure was used for mCherry and 1-s exposure for sfGFP. ImageJ software was employed for quantification to calculate fluorescence intensities in the red and green channels relative to that in a neighboring background region for each bacterium, and a GFP/mCherry ratio was generated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Scott Gray-Owen and members of his lab, in particular, Ryan Gaudet, for their generous donation of technical expertise and macrophage cell lines and use of their equipment.
W.W.N. was supported by an operating grant from the Canada Institutes for Health Research (MOP-86683) and a Natural Sciences and Engineering Research Council (NSERC) of Canada grant (RGPIN 386286-10). S.J.H. was supported by an NSERC Vanier Canada graduate scholarship.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hansen-Wester I, Hensel M. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect 3:549–559. doi: 10.1016/s1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- 2.Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella Typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A 83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäumler AJ, Tsolis RM, Ficht TA, Adams LG. 1998. Evolution of host adaptation in Salmonella enterica. Infect Immun 66:4579–4587. doi: 10.1128/IAI.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäumler AJ, Kusters JG, Stojiljkovic I, Heffron F. 1994. Salmonella Typhimurium loci involved in survival within macrophages. Infect Immun 62:1623–1630. doi: 10.1128/IAI.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spiga L, Winter MG, Furtado de Carvalho T, Zhu W, Hughes ER, Gillis CC, Behrendt CL, Kim J, Chessa D, Andrews-Polymenis HL, Beiting DP, Santos RL, Hooper LV, Winter SE. 2017. An oxidative central metabolism enables Salmonella to utilize microbiota-derived succinate. Cell Host Microbe 22:291.e6–301.e6. doi: 10.1016/j.chom.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordes T, Michelucci A, Hiller K. 2015. Itaconic acid: the surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annu Rev Nutr 35:451–473. doi: 10.1146/annurev-nutr-071714-034243. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z, Wang H, Wang Y, Ren Y, Wei D. 2018. Manufacturing multienzymatic complex reactors in vivo by self-assembly to improve the biosynthesis of itaconic acid in Escherichia coli. ACS Synth Biol 7:1244–1250. doi: 10.1021/acssynbio.8b00086. [DOI] [PubMed] [Google Scholar]
- 10.Zhao M, Lu X, Zong H, Li J, Zhuge B. 2018. Itaconic acid production in microorganisms. Biotechnol Lett 40:455–464. doi: 10.1007/s10529-017-2500-5. [DOI] [PubMed] [Google Scholar]
- 11.Hoyt JC, Robertson EF, Berlyn KA, Reeves HC. 1988. Escherichia coli isocitrate lyase: properties and comparisons. Biochim Biophys Acta 966:30–35. doi: 10.1016/0304-4165(88)90125-0. [DOI] [PubMed] [Google Scholar]
- 12.McFadden BA, Purohit S. 1977. Itaconate, an isocitrate lyase-directed inhibitor in Pseudomonas indigofera. J Bacteriol 131:136–144. doi: 10.1128/JB.131.1.136-144.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, Buttini M, Linster CL, Medina E, Balling R, Hiller K. 2013. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci U S A 110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strelko CL, Lu W, Dufort FJ, Seyfried TN, Chiles TC, Rabinowitz JD, Roberts MF. 2011. Itaconic acid is a mammalian metabolite induced during macrophage activation. J Am Chem Soc 133:16386–16389. doi: 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naujoks J, Tabeling C, Dill BD, Hoffmann C, Brown AS, Kunze M, Kempa S, Peter A, Mollenkopf H-J, Dorhoi A, Kershaw O, Gruber AD, Sander LE, Witzenrath M, Herold S, Nerlich A, Hocke AC, van Driel I, Suttorp N, Bedoui S, Hilbi H, Trost M, Opitz B. 2016. IFNs modify the proteome of Legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog 12:e1005408. doi: 10.1371/journal.ppat.1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, Lei H, Wu J, Li JV, Tang H, Wang Y. 2014. Systemic responses of BALB/c mice to Salmonella Typhimurium infection. J Proteome Res 13:4436–4445. doi: 10.1021/pr500770x. [DOI] [PubMed] [Google Scholar]
- 17.Sasikaran J, Ziemski M, Zadora PK, Fleig A, Berg IA. 2014. Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol 10:371–377. doi: 10.1038/nchembio.1482. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 19.Srikumar S, Kröger C, Hébrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron ADS, Hokamp K, Hinton J. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog 11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi L, Adkins JN, Coleman JR, Schepmoes AA, Dohnkova A, Mottaz HM, Norbeck AD, Purvine SO, Manes NP, Smallwood HS, Wang H, Forbes J, Gros P, Uzzau S, Rodland KD, Heffron F, Smith RD, Squier TC. 2006. Proteomic analysis of Salmonella enterica serovar Typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar Typhimurium inside macrophages. J Biol Chem 281:29131–29140. doi: 10.1074/jbc.M604640200. [DOI] [PubMed] [Google Scholar]
- 21.Santiviago CA, Reynolds MM, Porwollik S, Choi SH, Long F, Andrews-Polymenis HL, McClelland M. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog 5:e1000477. doi: 10.1371/journal.ppat.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haneda T, Ishii Y, Danbara H, Okada N. 2009. Genome-wide identification of novel genomic islands that contribute to Salmonella virulence in mouse systemic infection. FEMS Microbiol Lett 297:241–249. doi: 10.1111/j.1574-6968.2009.01686.x. [DOI] [PubMed] [Google Scholar]
- 23.Elder JR, Chiok KL, Paul NC, Haldorson G, Guard J, Shah DH. 2016. The Salmonella pathogenicity island 13 contributes to pathogenesis in streptomycin pre-treated mice but not in day-old chickens. Gut Pathog 8:16. doi: 10.1186/s13099-016-0098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espinoza RA, Silva-Valenzuela CA, Amaya FA, Urrutia IM, Contreras I, Santiviago CA. 2017. Differential roles for pathogenicity islands SPI-13 and SPI-8 in the interaction of Salmonella Enteritidis and Salmonella Typhi with murine and human macrophages. Biol Res 50:5. doi: 10.1186/s40659-017-0109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hersch SJ, Navarre WW. 6 May 2020. The Salmonella LysR family regulator, RipR, activates the SPI-13 encoded itaconate degradation cluster. BioRxiv doi: 10.1101/648865. [DOI] [PMC free article] [PubMed]
- 26.Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 27.Alpuche Aranda CM, Swanson JA, Loomis WP, Miller SI. 1992. Salmonella Typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci U S A 89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rathman M, Sjaastad MD, Falkow S. 1996. Acidification of phagosomes containing Salmonella Typhimurium in murine macrophages. Infect Immun 64:2765–2773. doi: 10.1128/IAI.64.7.2765-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakraborty S, Mizusaki H, Kenney LJ. 2015. A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol 13:e1002116. doi: 10.1371/journal.pbio.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E, Szpyt J, Runtsch MC, King MS, McGouran JF, Fischer R, Kessler BM, McGettrick AF, Hughes MM, Carroll RG, Booty LM, Knatko EV, Meakin PJ, Ashford MLJ, Modis LK, Brunori G, Sévin DC, Fallon PG, Caldwell ST, Kunji ERS, Chouchani ET, Frezza C, Dinkova-Kostova AT, Hartley RC, Murphy MP, O'Neill LA. 2018. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jha AK, Huang S-C, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ, Driggers EM, Artyomov MN. 2015. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Tallam A, Perumal TM, Antony PM, Jager C, Fritz JV, Vallar L, Balling R, Del Sol A, Michelucci A. 2016. Gene regulatory network inference of immunoresponsive Gene 1 (IRG1) identifies interferon regulatory factor 1 (IRF1) as its transcriptional regulator in mammalian macrophages. PLoS One 11:e0149050. doi: 10.1371/journal.pone.0149050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, Bouley DM, Vilches-Moure JG, Smith M, Sonnenburg JL, Bhatt AS, Huang KC, Monack D. 2018. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe 24:296.e7–307.e7. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricke SC. 2003. Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poult Sci 82:632–639. doi: 10.1093/ps/82.4.632. [DOI] [PubMed] [Google Scholar]
- 35.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O'Neill LAJ. 2013. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammerer F, Chang JH, Duncan D, Castaneda Ruiz A, Auclair K. 2016. Small molecule restores itaconate sensitivity in Salmonella enterica: a potential new approach to treating bacterial infections. Chembiochem 17:1513–1517. doi: 10.1002/cbic.201600078. [DOI] [PubMed] [Google Scholar]
- 37.Schmieger H. 1971. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet 110:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 38.Zou SB, Hersch SJ, Roy H, Wiggers JB, Leung AS, Buranyi S, Xie JL, Dare K, Ibba M, Navarre WW. 2012. Loss of elongation factor P disrupts bacterial outer membrane integrity. J Bacteriol 194:413–425. doi: 10.1128/JB.05864-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 41.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 42.Corcoran CP, Podkaminski D, Papenfort K, Urban JH, Hinton JCD, Vogel J. 2012. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol 84:428–445. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- 43.Urban J, Vogel J. 2009. A green fluorescent protein (GFP)-based plasmid system to study post-transcriptional control of gene expression in vivo. Methods Mol Biol 540:301–319. doi: 10.1007/978-1-59745-558-9_22. [DOI] [PubMed] [Google Scholar]
- 44.Cooper CA, Zhang K, Andres SN, Fang Y, Kaniuk NA, Hannemann M, Brumell JH, Foster LJ, Junop MS, Coombes BK. 2010. Structural and biochemical characterization of SrcA, a multi-cargo type III secretion chaperone in Salmonella required for pathogenic association with a host. PLoS Pathog 6:e1000751. doi: 10.1371/journal.ppat.1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coombes BK, Brown NF, Valdez Y, Brumell JH, Finlay BB. 2004. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J Biol Chem 279:49804–49815. doi: 10.1074/jbc.M404299200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.