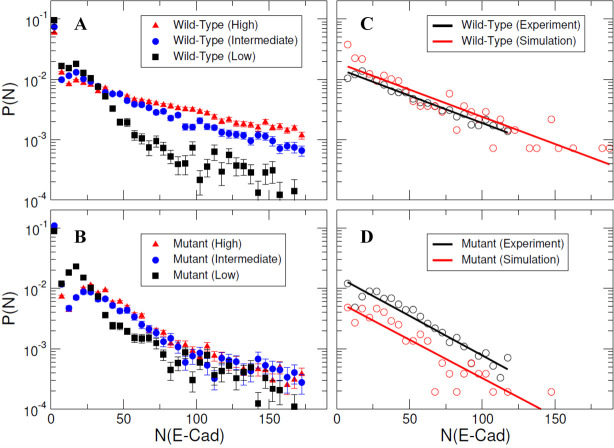
Figure 5. Specific and nonspecific interactions can cause E-cad clustering.
(A–B) Representative experimental cluster size probability distribution functions for wild-type and mutant E-cad at low, intermediate, and high surface coverages. Error bars correspond to the standard deviation of cluster size probability distribution functions calculated using 100 samples using a bootstrap method with replacement. (C–D) The comparison of experimental and simulated cluster size distributions for mutant and wild-type E-cad. The solid lines indicate the single exponential fitting.
Figure 5—figure supplement 1. Distributions of mutant E-cad cluster size for different combinations of nonspecific interaction on/off rate.
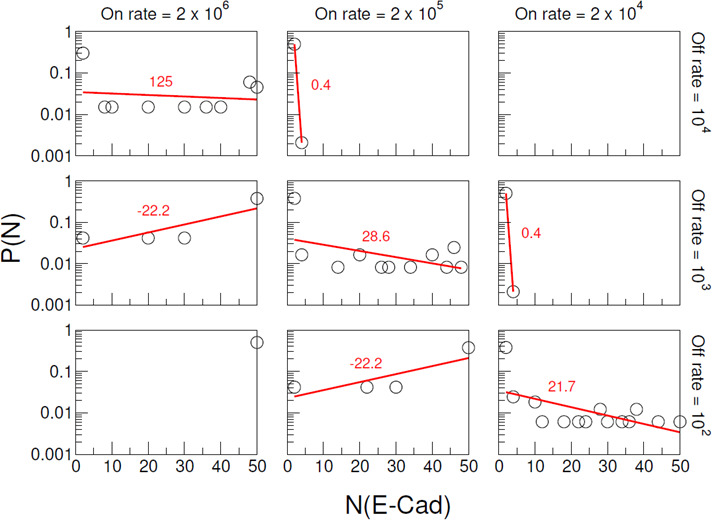
Figure 5—figure supplement 2. Distributions of wild-type E-cad cluster size for different combinations of specific interaction on/off rate.
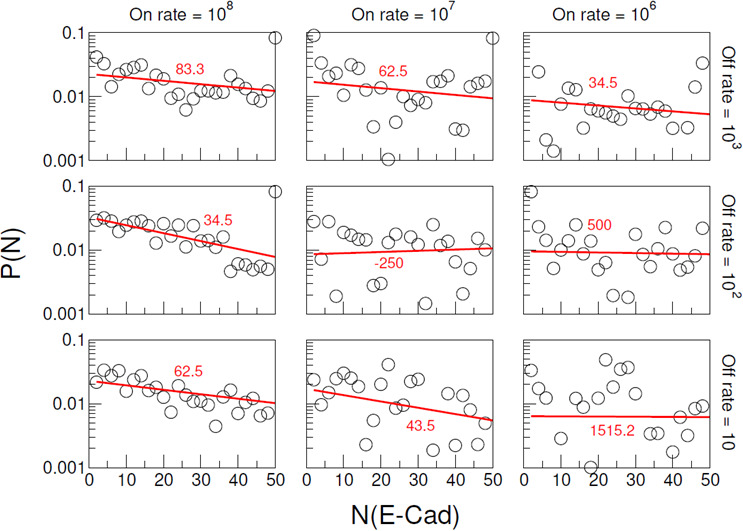
Figure 5—figure supplement 3. E-cad is primarily bound to a single lipid.
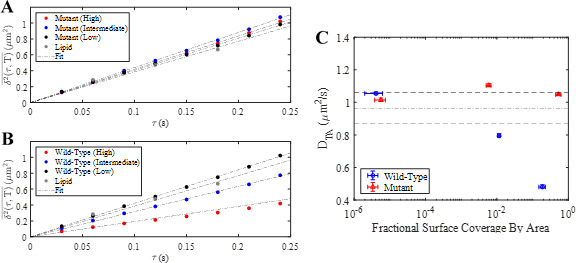
Figure 5—figure supplement 4. Trajectory averaged friction factor probability distributions for wild-type (top) and mutant (bottom) E-cad at high, intermediate, and low E-cad surface coverage.
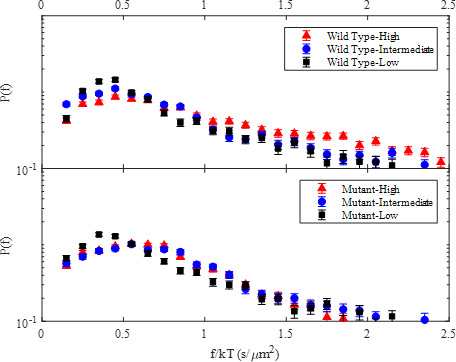
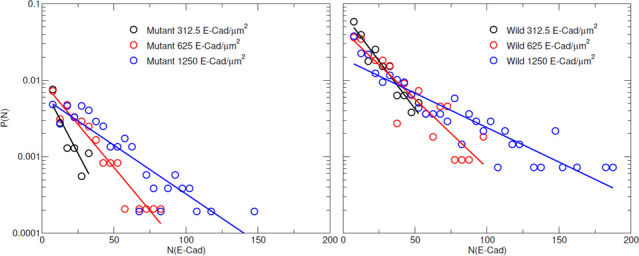
Figure 5—figure supplement 5. Distribution of cluster size for wild-type and mutant E-cad at a surface density of 312.5 E-cad/µm2, 625 E-cad/µm2, and 1,250 E-cad/µm2, respectively.