(
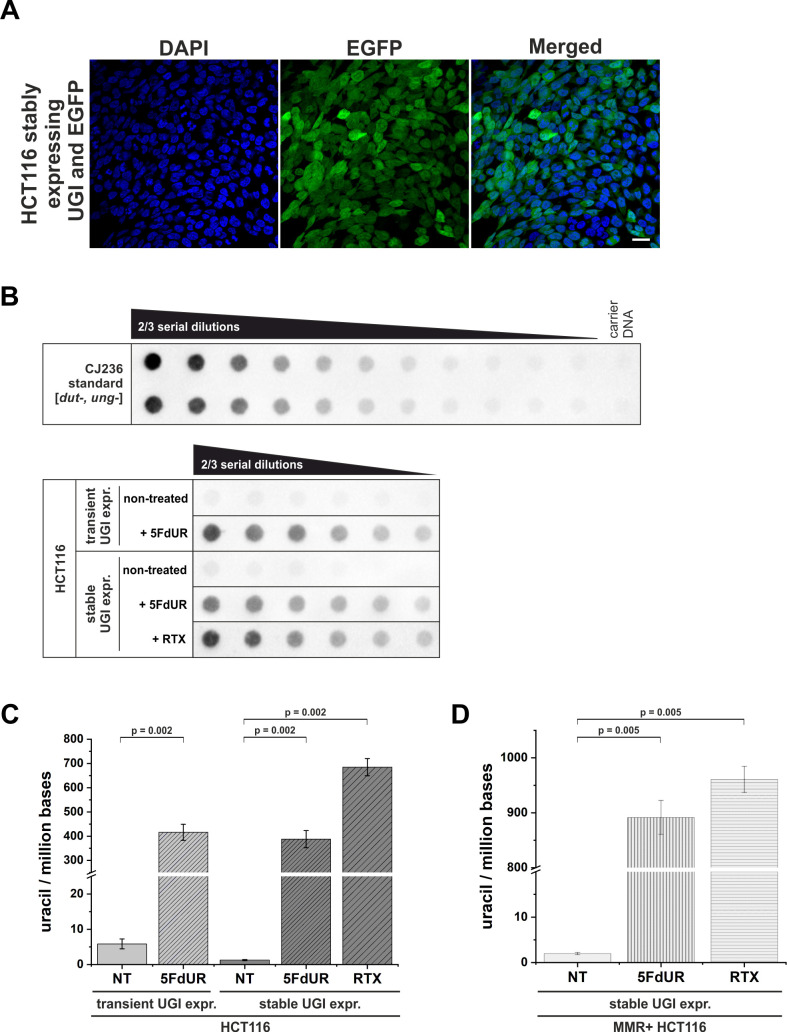
A) HCT116 cells stably expressing UGI along with EGFP following retroviral transduction. GFP positive cells were selected by fluorescence-activated cell sorting and cultured for further analysis. DAPI was used for DNA staining. Scale bar represents 20 µm. (
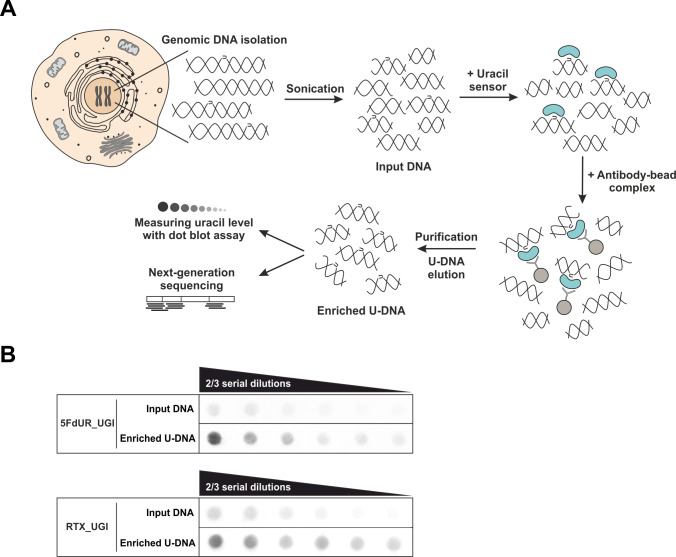
B) Dot blot assay for measuring genomic uracil levels (
Róna et al., 2016) of non-treated and drug (5FdUR or RTX) treated HCT116 cells either transiently or stably expressing UGI. Genomic DNA (8 ng) isolated from log-phase growing CJ236 [
dut-, ung-]
Escherichia coli strain was applied as uracil standard in a ½ dilution series (upper panel). Two-third dilution series from HCT116 samples started with 600 ng DNA for non-treated or 5 ng DNA for 5FdUR or RTX treated samples (lower panel). The dot blot image presented is a representative of six independent biological experiments. (
C) Bar graph shows the uracil moieties/million bases of each sample. Drug treatment led to significantly elevated uracil levels in HCT116 cells either transiently or stably expressing UGI (~400 uracil moieties/million for 5FdUR and ~700 uracil moieties/million for RTX) as compared to non-treated (NT) cells (~2–5 uracil moieties/million). Error bars indicate standard errors of the mean (SEM). Calculations were based on six independent datasets (n = 6). p=0.002. Source data are available in
Figure 1—figure supplement 1—source data 1. (
D) Bar graph shows the uracil moieties/million bases in MMR proficient HCT116 cells stably expressing UGI. Drug treatment resulted in even higher U-DNA content (~900 uracil moieties/million for 5FdUR and ~950 uracil moieties/million for RTX) as compared to MMR deficient cells. Error bars indicate standard errors of the mean (SEM). Calculations were based on six independent datasets (n = 6). p=0.005. Source data are available in
Figure 1—figure supplement 1—source data 1.