Figure 4. Characterization of U-DNA enrichment patterns.
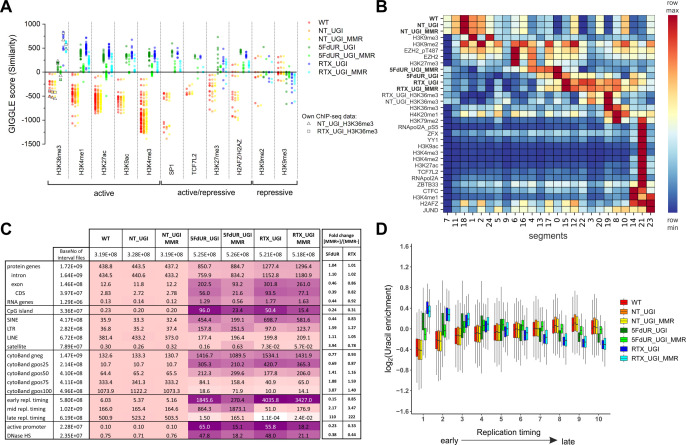
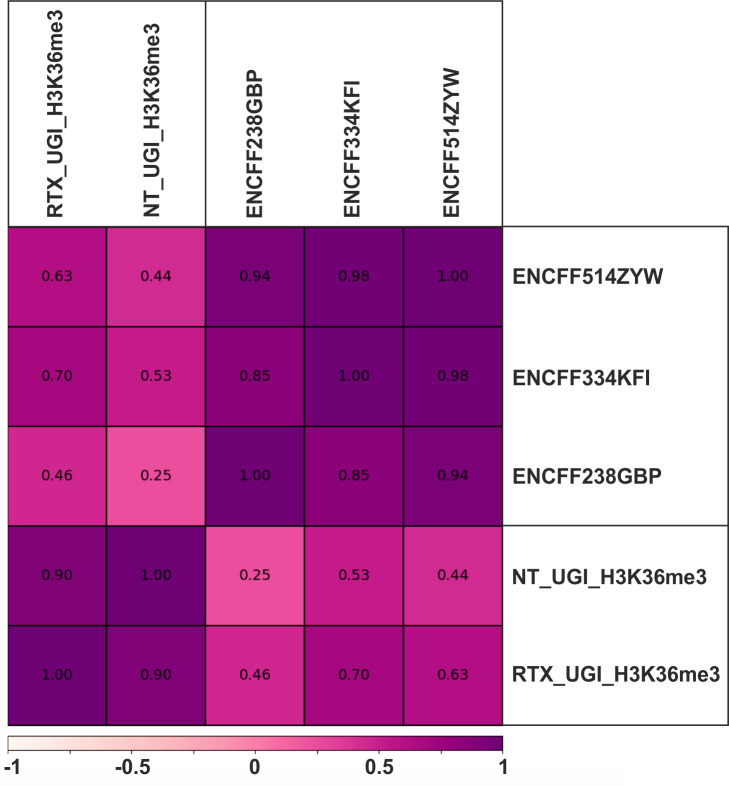
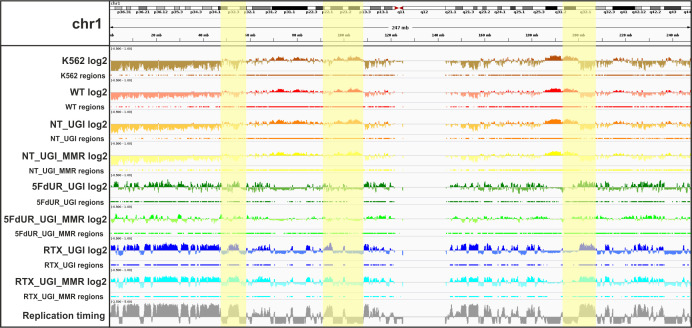
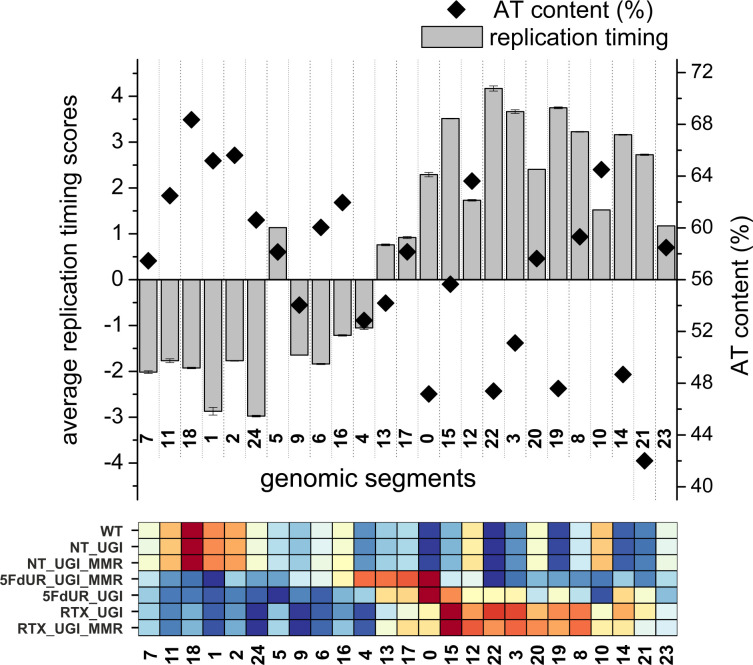
(A) GIGGLE search was performed with interval (bed) files of uracil enriched regions on a set of HCT116 related ChIP-seq and DIP-seq experimental data (for details see Supplementary file 3). Factors corresponding to the top 10 hits for each sample were selected. GIGGLE scores between all seven samples and all experiments corresponding to these factors were plotted excluding those, where data were not informative (data are found in Supplementary file 3-table 1). Source data are available in Figure 4—source data 1. Histone marks and the transcription factors, SP1 and TCF7L2 are categorized depending on their occurrence in transcriptionally active or repressive regions. Notably, some of them have plastic behavior allowing either transcriptionally active or repressive function. U-DNA-Seq samples are as follows: non-treated wild type (WT, red), non-treated UGI-expressing (NT_UGI, orange), 5FdUR treated UGI-expressing (5FdUR_UGI, green) and RTX treated UGI-expressing (RTX_UGI, blue) HCT116 cells, and their MMR proficient counterparts (NT_UGI_MMR, yellow; 5FdUR_UGI_MMR, light green; RTX_UGI_MMR, light blue). GIGGLE scores are also indicated for our own H3K36me3 ChIP-seq experiments (RTX_UGI sample: empty squares, NT_UGI sample: empty triangles). The tendencies are even more pronounced if the RTX treated U-DNA-Seq is compared with the RTX treated ChIP-seq or if the non-treated U-DNA-Seq is compared with the non-treated ChIP-seq data. (B) Genome segmentation analysis was performed on signal tracks of 22 ChIP-seq data available for HCT116 cells in the ENCODE database, on our own ChIP-seq data for H3K36me3, and on the seven U-DNA enrichment profiles (bold). The Segway train was performed with 25 labels and the corresponding genomic segments were identified with Segway annotate (Chan et al., 2018). The signal distribution data were calculated using Segtools (Buske et al., 2011), and plotted using python seaborn/matplotlib modules (Hunter, 2007). Source data are available in Figure 4—source data 2. Details including the applied command lines are provided in Supplementary file 3. The color-code is applied for each factor (rows) independently, from the minimum to the maximum value as indicated. (C) Correlation with genomic features. Interval (bed) files of genomic features were obtained from UCSC, Ensembl, and ReplicationDomain databases (for details see Supplementary file 4-table 1), and correlation with interval files of uracil regions were analyzed using bedtools annotate software (details are provided in Supplementary file 4). Numbers of overlapping base pairs were summarized for each pair of interval files, and scores were calculated according the formula: (baseNo_overlap/baseNo_sample_file) * (baseNo_overlap/baseNo_feature_file) * 10000. Heatmap was created based on fold increase of the scores compared to the corresponding WT scores. Sizes of interval files in number of base pairs are also given in the second column and the second line. Upon drug treatments, a clear shift from non-coding/heterochromatic/late replicated segments towards more active/coding/euchromatic/early replicated segments can be seen. CDS, coding sequence; SINE, short interspersed element; LTR, long terminal repeat; LINE, long interspersed element; cytoBand, cytogenetic chromosome band negatively (gneg) or positively (gpos) stained by Giemsa; repl. timing, replication timing; DNaseHS, DNase hypersensitive site. (D) Correlation analysis with replication timing. Replication timing data (bigWig files with 5000 bp binsize) specific for HCT116 were downloaded from ReplicationDomain database (Weddington et al., 2008). Data bins were distributed to 10 equal size groups according to replication timing from early to late. Then log2 uracil enrichment signals for these data bin groups were plotted for each sample using R (Supplementary file 5). Source data are available in Figure 4—source data 3.