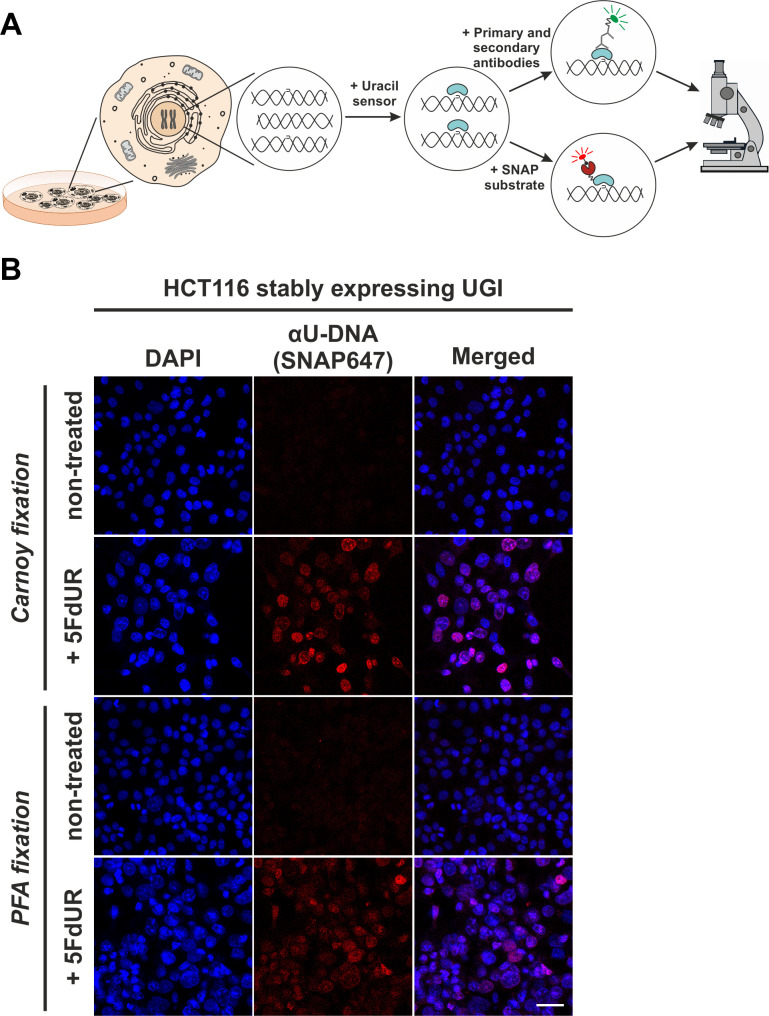
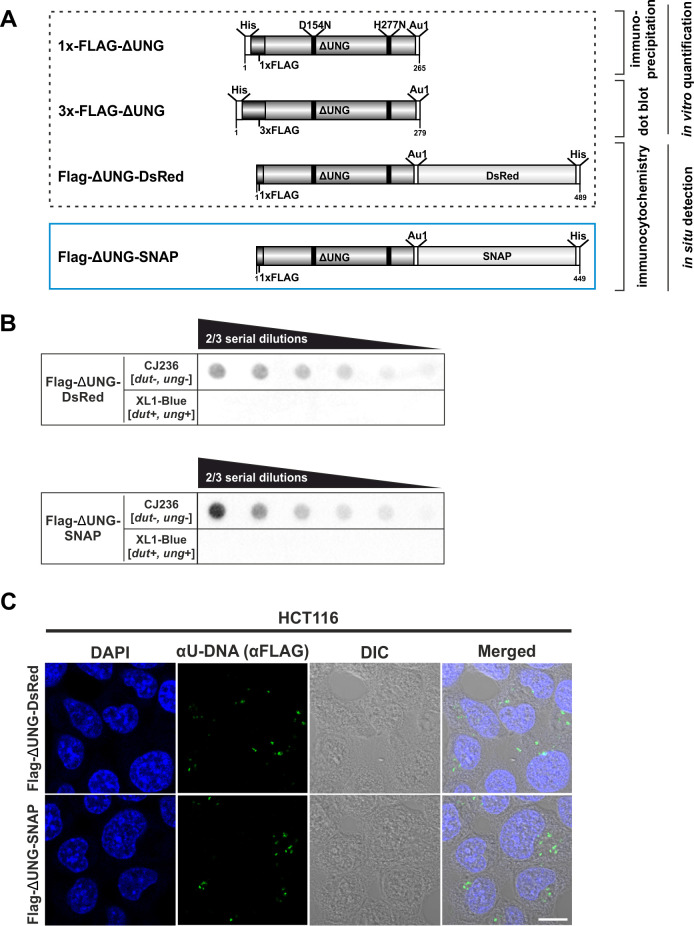
Figure 6. In situ detection of the cellular endogenous U-DNA content.
(A). Scheme represents that genomic uracil residues can be visualized in situ using our further developed U-DNA sensor construct via immunocytochemistry (through FLAG-tag) or directly via SNAP-tag chemistry. (B) HCT116 cells expressing UGI and treated with 5FdUR show efficient staining with the uracil sensor compared to non-treated cells, detected by confocal microscopy. Uracil residues are labelled by our FLAG-ΔUNG-SNAP sensor protein visualized by the SNAP647 substrate. DAPI was used for DNA counterstaining. Our optimized staining method is capable of comparable, specific uracil detection in HCT116 cells even with paraformaldehyde (PFA) fixation compared to the Carnoy fixation applied previously (Róna et al., 2016). Scale bar represents 40 µm. Note that the nuclei of the treated cells (5FdUR_UGI) are enlarged as compared to the non-treated ones (NT_UGI) presumably due to cell cycle arrest (Huehls et al., 2016; Yan et al., 2016).